Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus
- PMID: 16081957
- PMCID: PMC1233986
- DOI: 10.1128/JCM.43.8.4107-4111.2005
Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus
Abstract
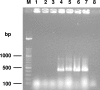
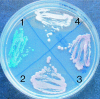
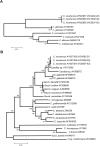
The new species Candida nivariensis, isolated from the clinical samples of three patients in Spain over a 3-year period, is presented here. This species can be easily differentiated from Candida glabrata, the closest genetic species, by different colony color on CHROMagar and by its ability to ferment trehalose. The analyses of the internal transcribed spacer region and the D1-D2 region of the 26S rRNA gene sequences support a new species designation.
Figures
References
-
- Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. - PubMed
-
- Casal, M., and M. J. Linares. 1981. Comparations of six media for production of chlamidospore by Candida albicans. Mycopathologia 76:125-128. - PubMed
-
- Feo, M., and A. De Pacheco. 1976. Candida albicans: milk in the production of chlamydospores. Rev. Latinoam. Microbiol. 18:23-24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

