Simultaneous detection of four human pathogenic microsporidian species from clinical samples by oligonucleotide microarray
- PMID: 16081959
- PMCID: PMC1233985
- DOI: 10.1128/JCM.43.8.4121-4128.2005
Simultaneous detection of four human pathogenic microsporidian species from clinical samples by oligonucleotide microarray
Abstract
Microsporidian species have been rapidly emerging as human enteric pathogens in immunocompromised and immunocompetent individuals in recent years. Routine diagnostic techniques for microsporidia in clinical laboratories are laborious and insensitive and tend to underestimate their presence. In most instances, they are unable to differentiate species of spores due to their small sizes and similar morphologies. In this study, we report the development of another protozoan oligonucleotide microarray assay for the simultaneous detection and identification to the species level of four major microsporidian species: Enterocytozoon bieneusi, Encephalitozoon cuniculi, Encephalitozoon hellem, and Encephalitozoon intestinalis. The 18S small-subunit rRNA gene was chosen as the amplification target, labeled with fluorescence dye, and hybridized to a series of species-specific oligonucleotide probes immobilized on a microchip. The specificity and sensitivity of the microarray were clearly demonstrated by the unique hybridization profiles exhibited by each species of microsporidian tested and its ability to detect as few as 10 spores. In order to assess the applicability of this microarray in a clinical setting, we conducted microarray assays of 20 fecal samples from AIDS patients. Twelve of these samples were positive for the presence of microsporidia and could be confidently identified; 11 of them were positive for more than one species. Our results suggested that this microarray-based approach represents an attractive diagnostic tool for high-throughput detection and identification of microsporidian species in clinical and epidemiological investigations.
Figures
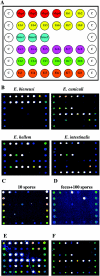
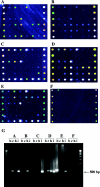
References
-
- Carville, A., K. Mansfield, G. Widmer, A. Lackner, D. Kotler, P. Wiest, T. Gumbo, S. Sarbah, and S. Tzipori. 1997. Development and application of genetic probes for detection of Enterocytozoon bieneusi in formalin-fixed stools and in intestinal biopsy specimens from infected patients. Clin. Diagn. Lab. Immunol. 4:405-408. - PMC - PubMed
-
- Charles, P. T., G. J. Vora, J. D. Andreadis, A. J. Fortney, C. E. Meador, C. S. Dulcey, and D. A. Stenger. 2003. Fabrication and surface characterization of DNA microarrays using amine- and thiol-terminated oligonucleotide probes. Langmuir 19:1586-1591.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

