The sentinel within: exploiting the immune system for cancer biomarkers
- PMID: 16083262
- PMCID: PMC2522321
- DOI: 10.1021/pr0500814
The sentinel within: exploiting the immune system for cancer biomarkers
Abstract
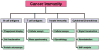
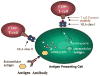
The release of proteins from tumors triggers an immune response in cancer patients. These tumor antigens arise from several mechanisms including tumor-specific alterations in protein expression, mutation, folding, degradation, or intracellular localization. Responses to most tumor antigens are rarely observed in healthy individuals, making the response itself a biomarker that betrays the presence of underlying cancer. Antibody immune responses show promise as clinical biomarkers because antibodies have long half-lives in serum, are easy to measure, and are stable in blood samples. However, our understanding of the specificity and the impact of the immune response in early stages of cancer is limited. The immune response to cancer, whether endogenous or driven by vaccines, involves highly specific T lymphocytes (which target tumor-derived peptides bound to self-MHC proteins) and B lymphocytes (which generate antibodies to tumor-derived proteins). T cell target antigens have been identified either by expression cloning from tumor cDNA libraries, or by prediction based on patterns of antigen expression ("reverse immunology"). B cell targets have been similarly identified using the antibodies in patient sera to screen cDNA libraries derived from tumor cell lines. This review focuses on the application of recent advances in proteomics for the identification of tumor antigens. These advances are opening the door for targeted vaccine development, and for using immune response signatures as biomarkers for cancer diagnosis and monitoring.
Figures


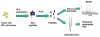
References
-
- Pavlenko M, Roos AK, Lundqvist A, Palmborg A, Miller AM, Ozenci V, Bergman B, Egevad L, Hellstrom M, Kiessling R, Masucci G, Wersall P, Nilsson S, Pisa P. A phase I trial of DNA vaccination with a plasmid expressing prostate-specific antigen in patients with hormone-refractory prostate cancer. Br J Cancer. 2004;91:688–694. - PMC - PubMed
-
- Vonderheide RH, Hahn WC, Schultze JL, Nadler LM. The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity. 1999;10:673–679. - PubMed
-
- Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res. 2000;60:1777–1788. - PubMed
-
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

