Initiation of DNA replication at the human beta-globin 3' enhancer
- PMID: 16085752
- PMCID: PMC1183104
- DOI: 10.1093/nar/gki747
Initiation of DNA replication at the human beta-globin 3' enhancer
Abstract
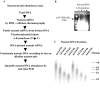
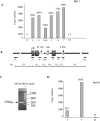
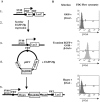
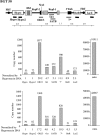
The origin of DNA replication in the human beta-globin gene contains an initiation region (IR) and two flanking auxiliary elements. Two replicator modules are located within the upstream auxiliary sequence and the IR core, but the functional sequences in the downstream auxiliary element are unknown. Here, we use a combination of benzoylated-naphthoylated DEAE (BND) cellulose purification and nascent strand abundance assays to show that replication initiation occurs at the beta-globin 3' enhancer on human chromosome 11 in the Hu11 hybrid murine erythroleukemia (MEL) cell line. To examine replicator function, 3' enhancer fragments were inserted into an ectopic site in MEL cells via an optimized FRT/EGFP-FLP integration system. These experiments demonstrate that the 1.6 kb downstream auxiliary element is a third replicator module called bGRep-E in erythroid cells. The minimal 260 bp 3' enhancer is required but not sufficient to initiate efficient replication, suggesting cooperation with adjacent sequences. The minimal 3' enhancer also cooperates with elements in an expressing HS3beta/gamma-globin construct to initiate replication. These data indicate that the beta-globin replicator has multiple initiation sites in three closely spaced replicator modules. We conclude that a mammalian enhancer can cooperate with adjacent sequences to create an efficient replicator module.
Figures



References
-
- Kitsberg D., Selig S., Keshet I., Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993;366:588–590. - PubMed
-
- Aladjem M.I., Groudine M., Brody L.L., Dieken E.S., Fournier R.E., Wahl G.M., Epner E.M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. - PubMed
-
- Aladjem M.I., Rodewald L.W., Kolman J.L., Wahl G.M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. - PubMed
-
- Pasceri P., Pannell D., Wu X., Ellis J. Full activity from human beta-globin locus control region transgenes requires 5′HS1, distal beta-globin promoter, and 3′ beta-globin sequences. Blood. 1998;92:653–663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

