Metabolism of the aliphatic nitramine 4-nitro-2,4-diazabutanal by Methylobacterium sp. strain JS178
- PMID: 16085803
- PMCID: PMC1183330
- DOI: 10.1128/AEM.71.8.4199-4202.2005
Metabolism of the aliphatic nitramine 4-nitro-2,4-diazabutanal by Methylobacterium sp. strain JS178
Abstract
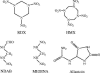
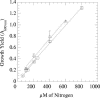
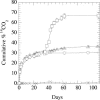
The aliphatic nitramine 4-nitro-2,4-diazabutanal (NDAB; C2H5N3O3) is a ring cleavage metabolite that accumulates during the aerobic degradation of the energetic compound hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by various Rhodococcus spp. NDAB is also produced during the alkaline hydrolysis of either RDX or octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX) and during the photolysis of RDX. Traces of NDAB were observed in a soil sampled from an ammunition-manufacturing facility contaminated with both HMX and RDX, suggesting natural attenuation. In this study, we report the isolation of a soil bacterium that is able to degrade NDAB under aerobic conditions. The isolate is a pink-pigmented facultative methylotroph affiliated with the genus Methylobacterium. The strain, named Methylobacterium sp. strain JS178, degrades NDAB as a sole nitrogen source, with concomitant growth and formation of 1 molar equivalent of nitrous oxide (N2O). Comparison of the growth yield of strain JS178 grown on NDAB, nitrite (NO2-), or ammonium (NH4+) as a nitrogen source revealed that 1 N equivalent is assimilated from each mole of NDAB, which completes the nitrogen mass balance. In radiotracer experiments, strain JS178 mineralized 1 C of the [14C]NDAB produced in situ from [14C]RDX by Rhodococcus sp. strain DN22. Studies on the regulation of NDAB degradation indicated that allantoin, an intermediate in the purine catabolic pathway and a central molecule in the storage and transport of nitrogen in plants, up-regulated the enzyme(s) involved in the degradation of the nitramine. The results reveal the potential for the sequential participation of rhodococci and methylobacteria to effect the complete degradation of RDX.
Figures

Similar articles
-
Biodegradation of the hexahydro-1,3,5-trinitro-1,3,5-triazine ring cleavage product 4-nitro-2,4-diazabutanal by Phanerochaete chrysosporium.Appl Environ Microbiol. 2004 Feb;70(2):1123-8. doi: 10.1128/AEM.70.2.1123-1128.2004. Appl Environ Microbiol. 2004. PMID: 14766596 Free PMC article.
-
Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a phytosymbiotic Methylobacterium sp. associated with poplar tissues (Populus deltoides x nigra DN34).Appl Environ Microbiol. 2004 Jan;70(1):508-17. doi: 10.1128/AEM.70.1.508-517.2004. Appl Environ Microbiol. 2004. PMID: 14711682 Free PMC article.
-
Phylogeny of cyclic nitramine-degrading psychrophilic bacteria in marine sediment and their potential role in the natural attenuation of explosives.FEMS Microbiol Ecol. 2004 Sep 1;49(3):349-57. doi: 10.1016/j.femsec.2004.04.008. FEMS Microbiol Ecol. 2004. PMID: 19712285
-
Microbial degradation and toxicity of hexahydro-1,3,5-trinitro-1,3,5-triazine.J Microbiol Biotechnol. 2012 Oct;22(10):1311-23. doi: 10.4014/jmb.1203.04002. J Microbiol Biotechnol. 2012. PMID: 23075780 Review.
-
Biodegradation of the cyclic nitramine explosives RDX, HMX, and CL-20.Appl Microbiol Biotechnol. 2006 Nov;73(2):274-90. doi: 10.1007/s00253-006-0588-y. Epub 2006 Oct 21. Appl Microbiol Biotechnol. 2006. PMID: 17058075 Review.
Cited by
-
Lateral transfer of genes for hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) degradation.Appl Environ Microbiol. 2009 May;75(10):3258-62. doi: 10.1128/AEM.02396-08. Epub 2009 Mar 6. Appl Environ Microbiol. 2009. PMID: 19270122 Free PMC article.
-
Microbially mediated biodegradation of hexahydro-1,3,5-trinitro-1,3,5- triazine by extracellular electron shuttling compounds.Appl Environ Microbiol. 2006 Sep;72(9):5933-41. doi: 10.1128/AEM.00660-06. Appl Environ Microbiol. 2006. PMID: 16957213 Free PMC article.
-
The Genome Analysis of Methylobacterium populi YC-XJ1 with Diverse Xenobiotics Biodegrading Capacity and Degradation Characteristics of Related Hydrolase.Int J Mol Sci. 2020 Jun 22;21(12):4436. doi: 10.3390/ijms21124436. Int J Mol Sci. 2020. PMID: 32580446 Free PMC article.
-
Biotransformation of N-nitrosodimethylamine by Pseudomonas mendocina KR1.Appl Environ Microbiol. 2006 Oct;72(10):6693-8. doi: 10.1128/AEM.01535-06. Epub 2006 Sep 1. Appl Environ Microbiol. 2006. PMID: 16950909 Free PMC article.
-
Activity assays of NnlA homologs suggest the natural product N-nitroglycine is degraded by diverse bacteria.Beilstein J Org Chem. 2024 Apr 17;20:830-840. doi: 10.3762/bjoc.20.75. eCollection 2024. Beilstein J Org Chem. 2024. PMID: 38655556 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. Wiley & Sons, New York, N.Y.
-
- Bhushan, B., S. Trott, J. C. Spain, A. Halasz, L. Paquet, and J. Hawari. 2003. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a rabbit liver cytochrome P450: insight into the mechanism of RDX biodegradation by Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 69:1347-1351. - PMC - PubMed
-
- Broder, M. F., and R. A. Westmoreland. 1998. An estimate of soils contaminated with secondary explosives. Final report. SFIM-AEC-ET-CR-98002. U.S. Army Environmental Center, Kansas City, Mo.
-
- Coleman, N. V., D. R. Nelson, and T. Duxbury. 1998. Aerobic degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) as a nitrogen source by a Rhodococcus sp., strain DN22. Soil Biol. Biochem. 30:1159-1167.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

