Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for Biofilms
- PMID: 16085832
- PMCID: PMC1183279
- DOI: 10.1128/AEM.71.8.4414-4426.2005
Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for Biofilms
Abstract
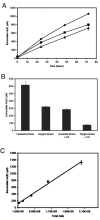
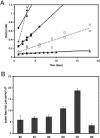
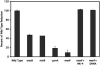
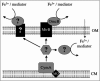
We developed a new method to measure iron reduction at a distance based on depositing Fe(III) (hydr)oxide within nanoporous glass beads. In this "Fe-bead" system, Shewanella oneidensis reduces at least 86.5% of the iron in the absence of direct contact. Biofilm formation accompanies Fe-bead reduction and is observable both macro- and microscopically. Fe-bead reduction is catalyzed by live cells adapted to anaerobic conditions, and maximal reduction rates require sustained protein synthesis. The amount of reactive ferric iron in the Fe-bead system is available in excess such that the rate of Fe-bead reduction is directly proportional to cell density; i.e., it is diffusion limited. Addition of either lysates prepared from anaerobic cells or exogenous electron shuttles stimulates Fe-bead reduction by S. oneidensis, but iron chelators or additional Fe(II) do not. Neither dissolved Fe(III) nor electron shuttling activity was detected in culture supernatants, implying that the mediator is retained within the biofilm matrix. Strains with mutations in omcB or mtrB show about 50% of the wild-type levels of reduction, while a cymA mutant shows less than 20% of the wild-type levels of reduction and a menF mutant shows insignificant reduction. The Fe-bead reduction defect of the menF mutant can be restored by addition of menaquinone, but menaquinone itself cannot stimulate Fe-bead reduction. Because the menF gene encodes the first committed step of menaquinone biosynthesis, no intermediates of the menaquinone biosynthetic pathway are used as diffusible mediators by this organism to promote iron reduction at a distance. CymA and menaquinone are required for both direct and indirect mineral reduction, whereas MtrB and OmcB contribute to but are not absolutely required for iron reduction at a distance.
Figures


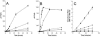
References
-
- Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. - PubMed
-
- Beveridge, T. J., S. A. Makin, J. L. Kadurugamuwa, and Z. S. Li. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20:291-303. - PubMed
-
- Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. ColladoVides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

