Host species-specific metabolic fingerprint database for enterococci and Escherichia coli and its application to identify sources of fecal contamination in surface waters
- PMID: 16085837
- PMCID: PMC1183341
- DOI: 10.1128/AEM.71.8.4461-4468.2005
Host species-specific metabolic fingerprint database for enterococci and Escherichia coli and its application to identify sources of fecal contamination in surface waters
Abstract
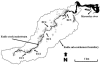
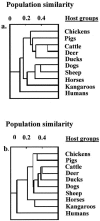
A metabolic fingerprint database of enterococci and Escherichia coli from 10 host groups of animals was developed to trace the sources of fecal contamination in surface waters. In all, 526 biochemical phenotypes (BPTs) of enterococci and 530 E. coli BPTs were obtained from 4,057 enterococci and 3,728 E. coli isolates tested. Of these, 231 Enterococcus BPTs and 257 E. coli BPTs were found in multiple host groups. The remaining 295 Enterococcus BPTs and 273 E. coli BPTs were unique to individual host groups. The database was used to trace the sources of fecal contamination in a local creek. The mean diversities (Di) of enterococci (Di = 0.76 +/- 0.05) and E. coli (Di = 0.88 +/- 0.04) were high (maximum 1) in water samples, indicating diverse sources of fecal contamination. Overall, 71% of BPTs of enterococci and 67% of E. coli BPTs from water samples were identified as human and animal sources. Altogether, 248 Enterococcus BPTs and 282 E. coli BPTs were found in water samples. Among enterococci, 26 (10%) BPTs were identical to those of humans and 152 BPTs (61%) were identical to those of animals (animal BPTs). Among E. coli isolates, 36 (13%) BPTs were identical to those of humans and 151 (54%) BPTs were identical to those of animals. Of the animal BPTs, 101 (66%) Enterococcus BPTs and 93 (62%) E. coli BPTs were also unique to individual animal groups. On the basis of these unique Enterococcus BPTs, chickens contributed 14% of contamination, followed by humans (10%), dogs (7%), and horses (6%). For E. coli, humans contributed 13% of contamination, followed by ducks (9%), cattle (7%), and chickens (6%). The developed metabolic fingerprint database was able to distinguish between human and animal sources as well as among animal species in the studied catchment.
Figures
References
-
- Abbot, S., B. Caughley, and G. Scott. 1993. Evaluation of Enterolert for the enumeration of enterococci in the marine environment. N. Z. J. Mar. Freshwater Res. 32:505-513.
-
- American Public Health Association. 1995. Standard method for the examination of water and wastewater, 19th ed. American Public Health Association, Inc., Washington, D.C.
-
- Anonymous. 1997. Water quality—detection and enumeration of coliform organisms, thermotolerant coliform organism and presumptive Escherichia coli. Part 1: membrane filtration method. International Organization for Standardization document 9308/1. ISO, Geneva, Switzerland.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

