Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture
- PMID: 16085862
- PMCID: PMC1183361
- DOI: 10.1128/AEM.71.8.4672-4678.2005
Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture
Abstract
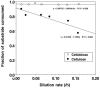
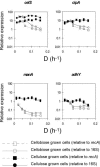
Clostridium thermocellum is a thermophilic, anaerobic, cellulolytic bacterium that produces ethanol and acetic acid as major fermentation end products. The effect of growth conditions on gene expression in C. thermocellum ATCC 27405 was studied using cells grown in continuous culture under cellobiose or cellulose limitation over a approximately 10-fold range of dilution rates (0.013 to 0.16 h(-1)). Fermentation product distribution displayed similar patterns in cellobiose- or cellulose-grown cultures, including substantial shifts in the proportion of ethanol and acetic acid with changes in growth rate. Expression of 17 genes involved or potentially involved in cellulose degradation, intracellular phosphorylation, catabolite repression, and fermentation end product formation was quantified by real-time PCR, with normalization to two calibrator genes (recA and the 16S rRNA gene) to determine relative expression. Thirteen genes displayed modest (fivefold or less) differences in expression with growth rate or substrate type: sdbA (cellulosomal scaffoldin-dockerin binding protein), cdp (cellodextrin phosphorylase), cbp (cellobiose phosphorylase), hydA (hydrogenase), ldh (lactate dehydrogenase), ack (acetate kinase), one putative type IV alcohol dehydrogenase, two putative cyclic AMP binding proteins, three putative Hpr-like proteins, and a putative Hpr serine kinase. By contrast, four genes displayed >10-fold-reduced levels of expression when grown on cellobiose at dilution rates of >0.05 h(-1): cipA (cellulosomal scaffolding protein), celS (exoglucanase), manA (mannanase), and a second type IV alcohol dehydrogenase. The data suggest that at least some cellulosomal components are transcriptionally regulated but that differences in expression with growth rate or among substrates do not directly account for observed changes in fermentation end product distribution.
Figures

Similar articles
-
Global gene expression patterns in Clostridium thermocellum as determined by microarray analysis of chemostat cultures on cellulose or cellobiose.Appl Environ Microbiol. 2011 Feb;77(4):1243-53. doi: 10.1128/AEM.02008-10. Epub 2010 Dec 17. Appl Environ Microbiol. 2011. PMID: 21169455 Free PMC article.
-
LacI Transcriptional Regulatory Networks in Clostridium thermocellum DSM1313.Appl Environ Microbiol. 2017 Feb 15;83(5):e02751-16. doi: 10.1128/AEM.02751-16. Print 2017 Mar 1. Appl Environ Microbiol. 2017. PMID: 28003194 Free PMC article.
-
Impact of pretreated Switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis.PLoS One. 2009;4(4):e5271. doi: 10.1371/journal.pone.0005271. Epub 2009 Apr 22. PLoS One. 2009. PMID: 19384422 Free PMC article.
-
Intracellular cellobiose metabolism and its applications in lignocellulose-based biorefineries.Bioresour Technol. 2017 Sep;239:496-506. doi: 10.1016/j.biortech.2017.05.001. Epub 2017 May 4. Bioresour Technol. 2017. PMID: 28535986 Review.
-
Phosphorylase-catalyzed bottom-up synthesis of short-chain soluble cello-oligosaccharides and property-tunable cellulosic materials.Biotechnol Adv. 2021 Nov 1;51:107633. doi: 10.1016/j.biotechadv.2020.107633. Epub 2020 Sep 20. Biotechnol Adv. 2021. PMID: 32966861 Review.
Cited by
-
Transcriptomic analysis of Clostridium thermocellum ATCC 27405 cellulose fermentation.BMC Microbiol. 2011 Jun 14;11:134. doi: 10.1186/1471-2180-11-134. BMC Microbiol. 2011. PMID: 21672225 Free PMC article.
-
Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression.BMC Microbiol. 2012 Sep 21;12:214. doi: 10.1186/1471-2180-12-214. BMC Microbiol. 2012. PMID: 22994686 Free PMC article.
-
Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis.BMC Genomics. 2014 Dec 4;15(1):1066. doi: 10.1186/1471-2164-15-1066. BMC Genomics. 2014. PMID: 25477200 Free PMC article.
-
Precise pretreatment of lignocellulose: relating substrate modification with subsequent hydrolysis and fermentation to products and by-products.Biotechnol Biofuels. 2017 Apr 11;10:88. doi: 10.1186/s13068-017-0775-3. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28400859 Free PMC article.
-
Differential β-glucosidase expression as a function of carbon source availability in Talaromyces amestolkiae: a genomic and proteomic approach.Biotechnol Biofuels. 2017 Jun 23;10:161. doi: 10.1186/s13068-017-0844-7. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28649280 Free PMC article.
References
-
- Alexander, J. K. 1972. Cellobiose phosphorylase from Clostridium thermocellum. Methods Enzymol. 28:944-948.
-
- Bothun, G. D., B. L. Knutson, J. A. Berberich, H. J. Strobel, and S. E. Nokes. 2004. Metabolic selectivity and growth of Clostridium thermocellum in continuous culture under elevated hydrostatic pressure. Appl. Microbiol. Biotechnol. 65:149-157. - PubMed
-
- Chen, J., and P. J. Weimer. 2001. Competition among three predominant ruminal cellulolytic bacteria in the absence or presence of non-cellulolytic bacteria. Microbiology 147:21-30. - PubMed
-
- Crosas, B., A. Allali-Hassani, S. E. Martinez, S. Martras, B. Persson, H. Jornvall, X. Pares, and J. Farres. 2000. Molecular basis for differential substrate specificity in class IV alcohol dehydrogenases. J. Biol. Chem. 275:25180-25187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

