Screening and identification of differentially expressed transcripts in circulating cells of prostate cancer patients using suppression subtractive hybridization
- PMID: 16086835
- PMCID: PMC1199617
- DOI: 10.1186/1476-4598-4-30
Screening and identification of differentially expressed transcripts in circulating cells of prostate cancer patients using suppression subtractive hybridization
Abstract
Background: Tumor metastasis and changes in host immunosurveillance are important components in cancer development. Tumor cell invasion into the bloodstream is an essential step for systemic metastasis. Currently, the detection of tumor cells in the circulation is mainly dependent upon the utilization of known epithelial cell markers. However, expression of these molecules is not limited to cancer patients; healthy people also have a small number of epithelial cells in their circulation. Utilizing these markers to detect circulating tumor cells (CTCs) cannot adequately explain the mechanisms of tumor cell survival or their development of metastatic potential in peripheral blood. The immune system can also evolve along with the cancer, actually promoting or selecting the outgrowth of tumor variants. Unfortunately, both metastasis and immunosurveillance remain mysterious and are debatable because we have yet to define the molecules that participate in these processes. We are interested in identifying the existence of expressed genes, or mRNA species, that are specifically associated with circulating cells of cancer-bearing patients using prostate cancer (PCa) as a model.
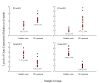
Results: We established two comprehensive subtracted cDNA libraries using a molecular technique called suppression subtractive hybridization. This technique selectively amplifies transcripts that are specifically expressed in circulating cells of either PCa patients or healthy men. Following sequencing reaction, we showed that 17 out of 23 (73.9%) sequenced clones did not match any mRNAs in the GenBank database. This result suggests that genes associated with alterations in circulating cells of cancer-bearing patients are largely unknown. Semi-quantitative RT-PCR confirmed that two genes are up-regulated in circulating cells of PCa patients, whereas another two genes are down-regulated in the same patients.
Conclusion: The comprehensive gene expression analysis is capable of identifying differentially expressed genes in circulating cells of healthy men and PCa patients. We did not attempt to enrich specific cell types in this study because phenotypes of CTCs and subsets of leukocytes participating in immunosurveillance remain largely unknown. Continuous studies of these differentially expressed genes will eventually lead us to understand the mechanisms involved in tumor metastasis and immune modulation during cancer development.
Figures


References
-
- Pantel K, Riethmuller G. Micrometastasis detection and treatment with monoclonal antibodies. Curr Top Microbiol Immunol. 1996;213:1–18. - PubMed
-
- Felton T, Harris GC, Pinder SE, Snead DR, Carter GI, Bell JA, Haines A, Kollias J, Robertson JF, Elston CW, Ellis IO. Identification of carcinoma cells in peripheral blood samples of patients with advanced breast carcinoma using RT-PCR amplification of CK7 and MUC1. Breast. 2004;13:35–41. doi: 10.1016/S0960-9776(03)00126-7. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

