The SET-domain protein superfamily: protein lysine methyltransferases
- PMID: 16086857
- PMCID: PMC1273623
- DOI: 10.1186/gb-2005-6-8-227
The SET-domain protein superfamily: protein lysine methyltransferases
Abstract
The SET-domain protein methyltransferase superfamily includes all but one of the proteins known to methylate histones on lysine. Histone methylation is important in the regulation of chromatin and gene expression.
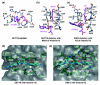
Figures


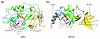
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

