Estimating brain functional connectivity with sparse multivariate autoregression
- PMID: 16087441
- PMCID: PMC1854937
- DOI: 10.1098/rstb.2005.1654
Estimating brain functional connectivity with sparse multivariate autoregression
Abstract
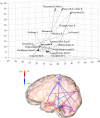
There is much current interest in identifying the anatomical and functional circuits that are the basis of the brain's computations, with hope that functional neuroimaging techniques will allow the in vivo study of these neural processes through the statistical analysis of the time-series they produce. Ideally, the use of techniques such as multivariate autoregressive (MAR) modelling should allow the identification of effective connectivity by combining graphical modelling methods with the concept of Granger causality. Unfortunately, current time-series methods perform well only for the case that the length of the time-series Nt is much larger than p, the number of brain sites studied, which is exactly the reverse of the situation in neuroimaging for which relatively short time-series are measured over thousands of voxels. Methods are introduced for dealing with this situation by using sparse MAR models. These can be estimated in a two-stage process involving (i) penalized regression and (ii) pruning of unlikely connections by means of the local false discovery rate developed by Efron. Extensive simulations were performed with idealized cortical networks having small world topologies and stable dynamics. These show that the detection efficiency of connections of the proposed procedure is quite high. Application of the method to real data was illustrated by the identification of neural circuitry related to emotional processing as measured by BOLD.
Figures









References
-
- Albert R, Barabasi A.L. Statistical mechanics of complex networks. Rev. Modern Phys. 2002;74:47–97.
-
- Baccala L.A, Sameshima K. Partial directed coherence: a new concept in neural structure determination. Biol. Cybern. 2001;84:463–474. - PubMed
-
- Bach F.R, Jordan M.I. Learning graphical models for stationary time series. IEEE Trans. Signal Proc. 2004;52:2189–2199.
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodological. 1995;57:289–300.
-
- Brillinger D.R, Bryant H.L, Segundo J.P. Identification of synaptic interactions. Biol. Cybern. 1976;22:213–228. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
