Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa
- PMID: 16087750
- PMCID: PMC1214527
- DOI: 10.1128/EC.4.8.1455-1464.2005
Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa
Abstract
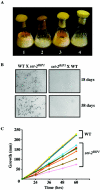
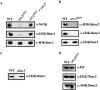
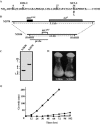
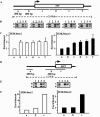
The SET domain is an evolutionarily conserved domain found predominantly in histone methyltransferases (HMTs). The Neurospora crassa genome includes nine SET domain genes (set-1 through set-9) in addition to dim-5, which encodes a histone H3 lysine 9 HMT required for DNA methylation. We demonstrate that Neurospora set-2 encodes a histone H3 lysine 36 (K36) methyltransferase and that it is essential for normal growth and development. We used repeat induced point mutation to make a set-2 mutant (set-2(RIP1)) with multiple nonsense mutations. Western analyses revealed that the mutant lacks SET-2 protein and K36 methylation. An amino-terminal fragment that includes the AWS, SET, and post-SET domains of SET-2 proved sufficient for K36 HMT activity in vitro. Nucleosomes were better substrates than free histones. The set-2(RIP1) mutant grows slowly, conidiates poorly, and is female sterile. Introducing the wild-type gene into the mutant complemented the defects, confirming that they resulted from loss of set-2 function. We replaced the wild-type histone H3 gene (hH3) with an allele producing a Lys to Leu substitution at position 36 and found that this hH3(K36L) mutant phenocopied the set-2(RIP1) mutant, confirming that the observed defects in growth and development result from inability to methylate K36 of H3. Finally, we used chromatin immunoprecipitation to demonstrate that actively transcribed genes in Neurospora crassa are enriched for H3 methylated at lysines 4 and 36. Taken together, our results suggest that methylation of K36 in Neurospora crassa is essential for normal growth and development.
Figures


References
-
- Bannister, A. J., R. Schneider, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2005. Spatial distribution of di-and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280:17732-17736. - PubMed
-
- Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

