DynGO: a tool for visualizing and mining of Gene Ontology and its associations
- PMID: 16091147
- PMCID: PMC1199584
- DOI: 10.1186/1471-2105-6-201
DynGO: a tool for visualizing and mining of Gene Ontology and its associations
Abstract
Background: A large volume of data and information about genes and gene products has been stored in various molecular biology databases. A major challenge for knowledge discovery using these databases is to identify related genes and gene products in disparate databases. The development of Gene Ontology (GO) as a common vocabulary for annotation allows integrated queries across multiple databases and identification of semantically related genes and gene products (i.e., genes and gene products that have similar GO annotations). Meanwhile, dozens of tools have been developed for browsing, mining or editing GO terms, their hierarchical relationships, or their "associated" genes and gene products (i.e., genes and gene products annotated with GO terms). Tools that allow users to directly search and inspect relations among all GO terms and their associated genes and gene products from multiple databases are needed.
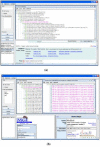
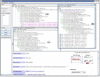
Results: We present a standalone package called DynGO, which provides several advanced functionalities in addition to the standard browsing capability of the official GO browsing tool (AmiGO). DynGO allows users to conduct batch retrieval of GO annotations for a list of genes and gene products, and semantic retrieval of genes and gene products sharing similar GO annotations. The result are shown in an association tree organized according to GO hierarchies and supported with many dynamic display options such as sorting tree nodes or changing orientation of the tree. For GO curators and frequent GO users, DynGO provides fast and convenient access to GO annotation data. DynGO is generally applicable to any data set where the records are annotated with GO terms, as illustrated by two examples.
Conclusion: We have presented a standalone package DynGO that provides functionalities to search and browse GO and its association databases as well as several additional functions such as batch retrieval and semantic retrieval. The complete documentation and software are freely available for download from the website http://biocreative.ifsm.umbc.edu/dyngo.
Figures






References
-
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–61. - PMC - PubMed
-
- Bult CJ, Blake JA, Richardson JE, Kadin JA, Eppig JT, Baldarelli RM, Barsanti K, Baya M, Beal JS, Boddy WJ, Bradt DW, Burkart DL, Butler NE, Campbell J, Corey R, Corbani LE, Cousins S, Dene H, Drabkin HJ, Frazer K, Garippa DM, Glass LH, Goldsmith CW, Grant PL, King BL, Lennon-Pierce M, Lewis J, Lu I, Lutz CM, Maltais LJ, McKenzie LM, Miers D, Modrusan D, Ni L, Ormsby JE, Qi D, Ramachandran S, Reddy TB, Reed DJ, Sinclair R, Shaw DR, Smith CL, Szauter P, Taylor B, Vanden Borre P, Walker M, Washburn L, Witham I, Winslow J, Zhu Y. The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res. 2004;32:D476–81. doi: 10.1093/nar/gkh125. - DOI - PMC - PubMed
-
- Twigger S, Lu J, Shimoyama M, Chen D, Pasko D, Long H, Ginster J, Chen CF, Nigam R, Kwitek A, Eppig J, Maltais L, Maglott D, Schuler G, Jacob H, Tonellato PJ. Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res. 2002;30:125–128. doi: 10.1093/nar/30.1.125. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

