Cross genome phylogenetic analysis of human and Drosophila G protein-coupled receptors: application to functional annotation of orphan receptors
- PMID: 16091152
- PMCID: PMC1192796
- DOI: 10.1186/1471-2164-6-106
Cross genome phylogenetic analysis of human and Drosophila G protein-coupled receptors: application to functional annotation of orphan receptors
Abstract
Background: The cell-membrane G-protein coupled receptors (GPCRs) are one of the largest known superfamilies and are the main focus of intense pharmaceutical research due to their key role in cell physiology and disease. A large number of putative GPCRs are 'orphans' with no identified natural ligands. The first step in understanding the function of orphan GPCRs is to identify their ligands. Phylogenetic clustering methods were used to elucidate the chemical nature of receptor ligands, which led to the identification of natural ligands for many orphan receptors. We have clustered human and Drosophila receptors with known ligands and orphans through cross genome phylogenetic analysis and hypothesized higher relationship of co-clustered members that would ease ligand identification, as related receptors share ligands with similar structure or class.
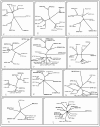
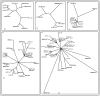
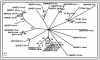
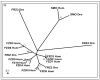
Results: Cross-genome phylogenetic analyses were performed to identify eight major groups of GPCRs dividing them into 32 clusters of 371 human and 113 Drosophila proteins (excluding olfactory, taste and gustatory receptors) and reveal unexpected levels of evolutionary conservation across human and Drosophila GPCRs. We also observe that members of human chemokine receptors, involved in immune response, and most of nucleotide-lipid receptors (except opsins) do not have counterparts in Drosophila. Similarly, a group of Drosophila GPCRs (methuselah receptors), associated in aging, is not present in humans.
Conclusion: Our analysis suggests ligand class association to 52 unknown Drosophila receptors and 95 unknown human GPCRs. A higher level of phylogenetic organization was revealed in which clusters with common domain architecture or cellular localization or ligand structure or chemistry or a shared function are evident across human and Drosophila genomes. Such analyses will prove valuable for identifying the natural ligands of Drosophila and human orphan receptors that can lead to a better understanding of physiological and pathological roles of these receptors.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

