siRNA-mediated off-target gene silencing triggered by a 7 nt complementation
- PMID: 16091630
- PMCID: PMC1184219
- DOI: 10.1093/nar/gki762
siRNA-mediated off-target gene silencing triggered by a 7 nt complementation
Abstract
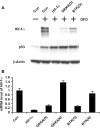
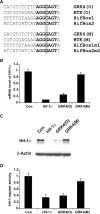
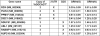
A growing body of evidence suggests that siRNA could generate off-target effects through different mechanisms. However, the full impact of off-target gene regulation on phenotypic induction and accordingly on data interpretation in the context of large-scale siRNA library screen has not been reported. Here we report on off-target gene silencing effects observed in a large-scale knockdown experiment designed to identify novel regulators of the HIF-1 pathway. All of the three 'top hits' from our screen have been demonstrated to result from off-target gene silencing. Two of the three 'siRNA hits' were found to directly trigger down-regulation of hif-1alpha mRNA through a 7 nt motif, AGGCAGT, that is present in both the hif-1alpha mRNA and the siRNAs. Further analysis revealed that the generation of off-target gene silencing via this 7 nt motif depends on the characteristics of the target mRNA, including the sequence context surrounding the complementary region, the position of the complementary region in the mRNA and the copy number of the complementary region. Interestingly, the off-target siRNA against hif-1alpha was also shown to trigger mRNA degradation with high probability of other genes that possess multiple copies of the AGGCAGT motif in the 3'-untranslated region. Lessons learned from this study will be a valuable asset to aid in designing siRNAs with more stringent target selectivity and improving 'hits-follow-up' strategies for future large-scale knockdown experiments.
Figures


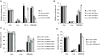
Similar articles
-
Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions.J Mol Biol. 2005 May 13;348(4):883-93. doi: 10.1016/j.jmb.2005.03.011. J Mol Biol. 2005. PMID: 15843020
-
[Effect of silencing HIF-1alpha by RNA interference on expression of vascular endothelial growth factor in osteosarcoma cell line SaOS-2 under hypoxia].Ai Zheng. 2005 May;24(5):531-5. Ai Zheng. 2005. PMID: 15890092 Chinese.
-
[The inhibition of VEGF expression induced by HIF-1alpha RNA interference].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2007 Oct;24(5):1133-6, 1166. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2007. PMID: 18027712 Chinese.
-
Is the Efficiency of RNA Silencing Evolutionarily Regulated?Int J Mol Sci. 2016 May 12;17(5):719. doi: 10.3390/ijms17050719. Int J Mol Sci. 2016. PMID: 27187367 Free PMC article. Review.
-
Precise and efficient siRNA design: a key point in competent gene silencing.Cancer Gene Ther. 2016 Apr;23(4):73-82. doi: 10.1038/cgt.2016.4. Epub 2016 Mar 18. Cancer Gene Ther. 2016. PMID: 26987292 Review.
Cited by
-
Single base mismatches in the mRNA target site allow specific seed region-mediated off-target binding of siRNA targeting human coagulation factor 7.RNA Biol. 2012 Jan;9(1):87-97. doi: 10.4161/rna.9.1.18121. Epub 2012 Jan 1. RNA Biol. 2012. PMID: 22258146 Free PMC article.
-
An off-target nucleostemin RNAi inhibits growth in human glioblastoma-derived cancer stem cells.PLoS One. 2011;6(12):e28753. doi: 10.1371/journal.pone.0028753. Epub 2011 Dec 12. PLoS One. 2011. PMID: 22174890 Free PMC article.
-
Selection of hyperfunctional siRNAs with improved potency and specificity.Nucleic Acids Res. 2009 Dec;37(22):e152. doi: 10.1093/nar/gkp864. Nucleic Acids Res. 2009. PMID: 19846596 Free PMC article.
-
Endoribonuclease-prepared short interfering RNAs induce effective and specific inhibition of human immunodeficiency virus type 1 replication.J Virol. 2007 Oct;81(19):10680-6. doi: 10.1128/JVI.00950-07. Epub 2007 Jul 25. J Virol. 2007. PMID: 17652404 Free PMC article.
-
Strand-specific 5'-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity.RNA. 2008 Feb;14(2):263-74. doi: 10.1261/rna.789808. Epub 2007 Dec 19. RNA. 2008. PMID: 18094121 Free PMC article.
References
-
- Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. - PubMed
-
- Aza-Blanc P., Cooper C.L., Wagner K., Batalov S., Deveraux Q.L., Cooke M.P. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell. 2003;12:627–637. - PubMed
-
- Brummelkamp T.R., Nijman S.M., Dirac A.M., Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

