Biophysical studies of a ruthenium(II) polypyridyl complex binding to DNA and RNA prove that nucleic acid structure has significant effects on binding behaviors
- PMID: 16091935
- PMCID: PMC7087908
- DOI: 10.1007/s00775-005-0007-3
Biophysical studies of a ruthenium(II) polypyridyl complex binding to DNA and RNA prove that nucleic acid structure has significant effects on binding behaviors
Abstract
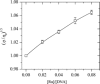
The interactions of a metal complex [Ru(phen)(2)PMIP](2+) {Ru=ruthenium, phen=1,10-phenanthroline, PMIP=2-(4-methylphenyl)imidazo[4,5-f]1,10-phenanthroline} with yeast tRNA and calf thymus DNA (CT DNA) have been investigated comparatively by UV-vis spectroscopy, fluorescence spectroscopy, viscosity measurements, isothermal titration calorimetry (ITC), as well as equilibrium dialysis and circular dichroism (CD). Spectroscopic studies together with ITC and viscosity measurements indicate that both binding modes of the Ru(II) polypyridyl complex to yeast tRNA and CT DNA are intercalation and yeast tRNA binding of the complex is stronger than CT DNA binding. ITC experiments show that the interaction of the complex with yeast tRNA is driven by a moderately favorable enthalpy decrease in combination with a moderately favorable entropy increase, while the binding of the complex to CT DNA is driven by a large favorable enthalpy decrease with a less favorable entropy increase. The results from equilibrium dialysis and CD suggest that both interactions are enantioselective and the Delta enantiomer of the complex may bind more favorably to both yeast tRNA and CT DNA than the Lambda enantiomer does, and that the complex is a better candidate for an enantioselective binder to yeast tRNA than to CT DNA. Taken together, these results indicate that the structures of nucleic acids have significant effects on the binding behaviors of metal complexes.
Figures





References
-
- Dupureur CM, Barton JK. Inorg Chem. 1997;36:33–43.
-
- Greguric I, Aldrich-Wright JR, Collins JG. J Am Chem Soc. 1997;119:3621–3622.
-
- Nair RB, Teng ES, Kirkland SL, Murphy CJ. Inorg Chem. 1998;37:139–141. - PubMed
-
- Liu JG, Ye BH, Li H, Ji LN, Li RH, Zhou JY. J Inorg Biochem. 1999;73:117–122.
-
- Onfelt B, Gostring L, Lincoln P, Nordén B, Onfelt A. Mutagenesis. 2002;17:317–320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

