Vitamin A and immune function: retinoic acid modulates population dynamics in antigen receptor and CD38-stimulated splenic B cells
- PMID: 16093312
- PMCID: PMC1242304
- DOI: 10.1073/pnas.0505018102
Vitamin A and immune function: retinoic acid modulates population dynamics in antigen receptor and CD38-stimulated splenic B cells
Abstract
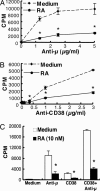
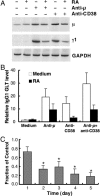
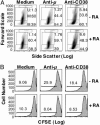
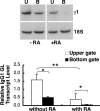
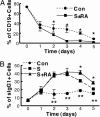
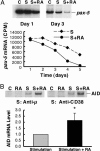
Vitamin A and its active metabolite, all-trans retinoic acid (RA), regulate the antibody response in vivo, although the underlying mechanisms are not well understood. We have investigated the regulation by RA of B cell population dynamics and Ig gene expression in purified splenic mouse B cells stimulated through the B cell antigen receptor (BCR) and/or CD38, a BCR coreceptor. After ligation of the BCR and/or CD38, B cells became more heterogeneous in size. RA substantially restrained this change, concomitant with inhibition of cell proliferation. To examine B cell heterogeneity more closely, we categorized stimulated B cells by size (forward angle light scatter) and determined cell division dynamics, germ-line Ig heavy chain gene transcription and surface IgG1 (sIgG1) expression. Flow cytometric analysis of carboxyfluorescein diacetate succinimidyl ester-labeled B cells costained for sIgG1 showed that the more proliferative groups of B cells were smaller, whereas cells expressing more sIgG1 were larger. RA enriched the latter population, whereas cell division frequency in general and the number of smaller B cells that had undergone division cycles were reduced. Although RA significantly inhibited Ig germ-line transcript levels in the total B cell population, CD19(-)IgG1(+) B cells, which represent a more differentiated phenotype, were enriched. Furthermore, pax-5 mRNA was decreased and activation-induced cytidine deaminase mRNA was increased in RA-treated stimulated B cells. Thus, RA regulated factors known to be required for Ig class switch recombination and modulated the population dynamics of ligation-stimulated B cells, while promoting the progression of a fraction of B cells into differentiated sIgG-expressing cells.
Figures


Comment in
-
Profile of A. Catharine Ross.Proc Natl Acad Sci U S A. 2005 Oct 4;102(40):14139-41. doi: 10.1073/pnas.0506178102. Epub 2005 Sep 26. Proc Natl Acad Sci U S A. 2005. PMID: 16186491 Free PMC article. No abstract available.
References
-
- Ross, A. C., Zolfaghari, R. & Weisz, J. (2001) Curr. Opin. Gastroenterol. 17, 184-192. - PubMed
-
- Reifen, R. (2002) Proc. Nutr. Soc. 61, 397-400. - PubMed
-
- Semba, R. D. (1999) Proc. Nutr. Soc. 58, 719-727. - PubMed
-
- Semba, R. D., Ndugwa, C., Perry, R. T., Clark, T. D., Jackson, J. B., Melikian, G., Tielsch, J. & Mmiro, F. (2005) Nutrition 21, 25-31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

