Rap1 prevents telomere fusions by nonhomologous end joining
- PMID: 16096640
- PMCID: PMC1201357
- DOI: 10.1038/sj.emboj.7600778
Rap1 prevents telomere fusions by nonhomologous end joining
Abstract
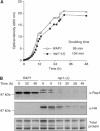
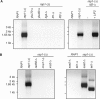
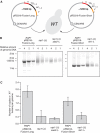
Telomeres protect chromosomes from end-to-end fusions. In yeast Saccharomyces cerevisiae, the protein Rap1 directly binds telomeric DNA. Here, we use a new conditional allele of RAP1 and show that Rap1 loss results in frequent fusions between telomeres. Analysis of the fusion point with restriction enzymes indicates that fusions occur between telomeres of near wild-type length. Telomere fusions are not observed in cells lacking factors required for nonhomologous end joining (NHEJ), including Lig4 (ligase IV), KU and the Mre11 complex. SAE2 and TEL1 do not affect the frequency of fusions. Together, these results show that Rap1 is essential to block NHEJ between telomeres. Since the presence of Rap1 at telomeres has been conserved through evolution, the establishment of NHEJ suppression by Rap1 could be universal.
Figures

References
-
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

