Towards an understanding of the structural molecular mechanism of beta(2)-microglobulin amyloid formation in vitro
- PMID: 16099226
- PMCID: PMC7617713
- DOI: 10.1016/j.bbapap.2005.07.006
Towards an understanding of the structural molecular mechanism of beta(2)-microglobulin amyloid formation in vitro
Abstract
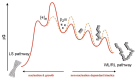
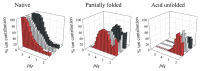
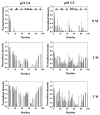
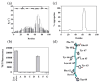
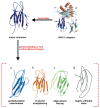
Deriving a complete understanding of protein self-association into amyloid fibrils across multiple distance and time scales is an enormous challenge. At small length scales, a detailed description of the partially folded protein ensemble that participates in self-assembly remains obscure. At larger length scales, amyloid fibrils are often heterogeneous, can form along multiple pathways, and are further complicated by phenomena such as phase-separation. Over the last 5 years, we have used an array of biophysical approaches in order to elucidate the structural and molecular mechanism of amyloid fibril formation, focusing on the all beta-sheet protein, beta(2)-microglubulin (beta(2)m). This protein forms amyloid deposits in the human disease 'dialysis-related amyloidosis' (DRA). We have shown that under acidic conditions beta(2)m rapidly associates in vitro to form amyloid-like fibrils that have different morphological properties, but which contain an underpinning cross-beta structure. In this review, we discuss our current knowledge of the structure of these fibrils, as well as the structural, kinetic and thermodynamic relationship between fibrils with different morphologies. The results provide some of the first insights into the shape of the self-assembly free-energy landscape for this protein and highlight the parallel nature of the assembly process. We include a detailed description of the structure and dynamics of partially folded and acid unfolded species of beta(2)m using NMR, and highlight regions thought to be important in early self-association events. Finally, we discuss briefly how knowledge of assembly mechanisms in vitro can be used to inform the design of therapeutic strategies for this, and other amyloid disorders, and we speculate on how the increasing power of biophysical approaches may lead to a fuller description of protein self-assembly into amyloid in the future.
Figures




References
-
- Gejyo F, Yamada T, Odani S, Nakagawa Y, Arakawa M, Kunitomo T, Kataoka H, Suzuki M, Hirasawa Y, Shirahama T, Cohen AS, et al. A new form of amyloid protein associated with chronic hemodialysis was identified as β2-microglobulin. Biochem Biophys Res Commun. 1985;129:701–706. - PubMed
-
- Floege J, Ketteler M. β2-microglobulin-derived amyloidosis: an update. Kidney Int. 2001;59:S164–S171. - PubMed
-
- Campistol JM, Bernard D, Papastoitsis G, Sole M, Kasirsky J, Skinner M. Polymerization of normal and intact β2-microglobulin as the amyloidogenic protein in dialysis-amyloidosis. Kidney Int. 1996;50:1262–1267. - PubMed
-
- Linke RP, Hampl H, Lobeck H, Ritz E, Bommer J, Waldherr R, Eulitz M. Lysine-specific cleavage of β2-microglobulin in amyloid deposits associated with hemodialysis. Kidney Int. 1989;36:675–681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

