Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription
- PMID: 16100256
- PMCID: PMC1366842
- DOI: 10.1529/biophysj.105.065326
Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription
Abstract
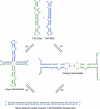
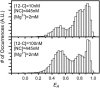
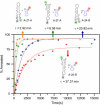
The minus-strand transfer step of HIV-1 reverse transcription is chaperoned by the nucleocapsid protein (NC), which has been shown to facilitate the annealing between the transactivation response element (TAR) RNA and complementary TAR DNA stem-loop structures. In this work, potential intermediates in the mechanism of NC-chaperoned TAR DNA/TAR RNA annealing have been examined using single-molecule fluorescence resonance energy transfer. The interaction between TAR DNA and various DNA oligonucleotides designed to mimic the initial annealing step was monitored to capture potential intermediates along the reaction pathway. Two possible mechanisms of annealing were examined, namely nucleation through the 3'/5' termini, termed the "zipper" complex, or nucleation through the hairpin loops in a "kissing" complex. Intermediates associated with both mechanisms were observed in the presence of NC, and the kinetics of formation of these intermediates were also measured. Thus, the single-molecule experiments support the notion that NC-assisted annealing of TAR DNA:TAR RNA may occur through multiple pathways.
Figures





Similar articles
-
Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein.J Mol Biol. 2006 Oct 13;363(1):244-61. doi: 10.1016/j.jmb.2006.08.039. Epub 2006 Aug 22. J Mol Biol. 2006. PMID: 16962137
-
Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription.Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5261-7. doi: 10.1073/pnas.0700166104. Epub 2007 Mar 19. Proc Natl Acad Sci U S A. 2007. PMID: 17372205 Free PMC article.
-
Probing nucleation, reverse annealing, and chaperone function along the reaction path of HIV-1 single-strand transfer.Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12651-6. doi: 10.1073/pnas.0700350104. Epub 2007 Jun 19. Proc Natl Acad Sci U S A. 2007. PMID: 17578926 Free PMC article.
-
Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism.Prog Nucleic Acid Res Mol Biol. 2005;80:217-86. doi: 10.1016/S0079-6603(05)80006-6. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164976 Review. No abstract available.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
Cited by
-
Single-molecule spectroscopic study of dynamic nanoscale DNA bending behavior of HIV-1 nucleocapsid protein.J Phys Chem B. 2013 Apr 25;117(16):4183-96. doi: 10.1021/jp3018259. Epub 2012 May 16. J Phys Chem B. 2013. PMID: 22591315 Free PMC article.
-
The HIV-1 nucleocapsid chaperone protein forms locally compacted globules on long double-stranded DNA.Nucleic Acids Res. 2021 May 7;49(8):4550-4563. doi: 10.1093/nar/gkab236. Nucleic Acids Res. 2021. PMID: 33872352 Free PMC article.
-
Probing dynamics of HIV-1 nucleocapsid protein/target hexanucleotide complexes by 2-aminopurine.Nucleic Acids Res. 2008 Feb;36(3):885-96. doi: 10.1093/nar/gkm1109. Epub 2007 Dec 17. Nucleic Acids Res. 2008. PMID: 18086707 Free PMC article.
-
Sensing peptide-oligonucleotide interactions by a two-color fluorescence label: application to the HIV-1 nucleocapsid protein.Nucleic Acids Res. 2009 Feb;37(3):e25. doi: 10.1093/nar/gkn1083. Epub 2009 Jan 16. Nucleic Acids Res. 2009. PMID: 19151084 Free PMC article.
-
Covalent protein-oligonucleotide conjugates by copper-free click reaction.Bioorg Med Chem. 2012 Jul 15;20(14):4532-9. doi: 10.1016/j.bmc.2012.05.017. Epub 2012 May 17. Bioorg Med Chem. 2012. PMID: 22682299 Free PMC article.
References
-
- Henderson, L. E., T. D. Copeland, R. C. Sowder, G. W. Smythers, and S. Oroszlan. 1981. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 256:8400–8406. - PubMed
-
- Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science. 232:485–487. - PubMed
-
- Levin, J. G., J. Guo, I. Rouzina, and K. Musier-Forsyth. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

