Compaction kinetics on single DNAs: purified nucleosome reconstitution systems versus crude extract
- PMID: 16100259
- PMCID: PMC1366857
- DOI: 10.1529/biophysj.105.062786
Compaction kinetics on single DNAs: purified nucleosome reconstitution systems versus crude extract
Abstract
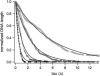
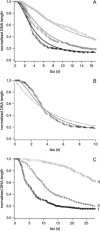
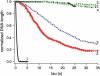
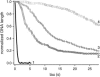
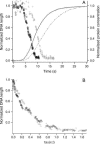
Kinetics of compaction on single DNA molecules are studied by fluorescence videomicroscopy in the presence of 1), Xenopus egg extracts and 2), purified nucleosome reconstitution systems using a combination of histones with either the histone chaperone Nucleosome Assembly Protein (NAP-1) or negatively charged macromolecules such as polyglutamic acid and RNA. The comparison shows that the compaction rates can differ by a factor of up to 1000 for the same amount of histones, depending on the system used and on the presence of histone tails, which can be subjected to post-translational modifications. Reactions with purified reconstitution systems follow a slow and sequential mechanism, compatible with the deposition of one (H3-H4)(2) tetramer followed by two (H2A-H2B) dimers. Addition of the histone chaperone NAP-1 increases both the rate of the reaction and the packing ratio of the final product. These stimulatory effects cannot be obtained with polyglutamic acid or RNA, suggesting that yNAP-1 impact on the reaction cannot simply be explained in terms of charge screening. Faster compaction kinetics and higher packing ratios are reproducibly reached with extracts, indicating a role of additional components present in this system. Data are discussed and models proposed to account for the kinetics obtained in our single-molecule assay.
Figures

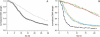
Similar articles
-
Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs.Proc Natl Acad Sci U S A. 2005 Jun 7;102(23):8210-5. doi: 10.1073/pnas.0500822102. Epub 2005 May 31. Proc Natl Acad Sci U S A. 2005. PMID: 15928086 Free PMC article.
-
Histone release during transcription: NAP1 forms a complex with H2A and H2B and facilitates a topologically dependent release of H3 and H4 from the nucleosome.Biochemistry. 2004 Mar 9;43(9):2359-72. doi: 10.1021/bi035737q. Biochemistry. 2004. PMID: 14992573
-
Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I.Mol Cell Biol. 2005 Dec;25(23):10639-51. doi: 10.1128/MCB.25.23.10639-10651.2005. Mol Cell Biol. 2005. PMID: 16287874 Free PMC article.
-
p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone.Genes Dev. 2000 Aug 1;14(15):1899-907. Genes Dev. 2000. PMID: 10921904 Free PMC article.
-
Current progress on structural studies of nucleosomes containing histone H3 variants.Curr Opin Struct Biol. 2013 Feb;23(1):109-15. doi: 10.1016/j.sbi.2012.10.009. Epub 2012 Dec 22. Curr Opin Struct Biol. 2013. PMID: 23265997 Review.
Cited by
-
Histone chaperone-mediated nucleosome assembly process.PLoS One. 2015 Jan 22;10(1):e0115007. doi: 10.1371/journal.pone.0115007. eCollection 2015. PLoS One. 2015. PMID: 25611318 Free PMC article.
-
A Quantitative Proteomic Analysis of In Vitro Assembled Chromatin.Mol Cell Proteomics. 2016 Mar;15(3):945-59. doi: 10.1074/mcp.M115.053553. Epub 2016 Jan 25. Mol Cell Proteomics. 2016. PMID: 26811354 Free PMC article.
-
Chromatin fiber dynamics under tension and torsion.Int J Mol Sci. 2010 Apr 12;11(4):1557-79. doi: 10.3390/ijms11041557. Int J Mol Sci. 2010. PMID: 20480035 Free PMC article. Review.
-
PIKK-dependent phosphorylation of Mre11 induces MRN complex inactivation by disassembly from chromatin.DNA Repair (Amst). 2009 Nov 2;8(11):1311-20. doi: 10.1016/j.dnarep.2009.07.006. Epub 2009 Aug 25. DNA Repair (Amst). 2009. PMID: 19709933 Free PMC article.
-
Histone H1 compacts DNA under force and during chromatin assembly.Mol Biol Cell. 2012 Dec;23(24):4864-71. doi: 10.1091/mbc.E12-07-0518. Epub 2012 Oct 24. Mol Biol Cell. 2012. PMID: 23097493 Free PMC article.
References
-
- Van Holde, K. E. 1988. Chromatin. Springer, New York.
-
- Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science. 293:1074–1080. - PubMed
-
- Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature. 403:41–45. - PubMed
-
- Carruthers, L. M., and J. C. Hansen. 2000. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275:37285–37290. - PubMed
-
- Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327:85–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

