Ligand binding modulates the mechanical stability of dihydrofolate reductase
- PMID: 16100277
- PMCID: PMC1366830
- DOI: 10.1529/biophysj.105.062034
Ligand binding modulates the mechanical stability of dihydrofolate reductase
Abstract
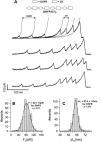
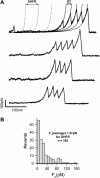
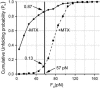
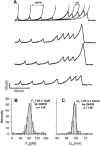
We use single-molecule force spectroscopy to demonstrate that the mechanical stability of the enzyme dihydrofolate reductase (DHFR) is modulated by ligand binding. In the absence of bound ligands, DHFR extends at very low forces, averaging 27 pN, without any characteristic mechanical fingerprint. By contrast, in the presence of micromolar concentrations of the ligands methotrexate, nicotinamide adenine dihydrogen phosphate, or dihydrofolate, much higher forces are required (82 +/- 18 pN, 98 +/- 15 pN, and 83 +/- 16 pN, respectively) and a characteristic fingerprint is observed in the force-extension curves. The increased mechanical stability triggered by these ligands is not additive. Our results explain the large reduction in the degradation rate of DHFR, in the presence of its ligands. Our observations support the view that the rate-limiting step in protein degradation by adenosine triphosphate-dependent proteases is the mechanical unfolding of the target protein.
Figures


Comment in
-
Fingerprinting DHFR in single-molecule AFM studies.Biophys J. 2006 Sep 1;91(5):2009-10, discussion 2011-2. doi: 10.1529/biophysj.106.085126. Epub 2006 Jun 16. Biophys J. 2006. PMID: 16782796 Free PMC article. No abstract available.
References
-
- Schnell, J. R., H. J. Dyson, and P. E. Wright. 2004. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 33:119–140. - PubMed
-
- Benkovic, S. J., and S. Hammes-Schiffer. 2003. A perspective on enzyme catalysis. Science. 301:1196–1202. - PubMed
-
- Pineda, P., A. Kanter, R. S. McIvor, S. J. Benkovic, A. Rosowsky, and C. R. Wagner. 2003. Dihydrofolate reductase mutant with exceptional resistance to methotrexate but not to trimetrexate. J. Med. Chem. 46:2816–2818. - PubMed
-
- Davies, J. F., T. J. Delcamp, N. J. Prendergast, V. A. Ashford, J. H. Freisheim, and J. Kraut. 1990. Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5-deazafolate. Biochemistry. 29:9467–9479. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

