Schoenheimer effect explained--feedback regulation of cholesterol synthesis in mice mediated by Insig proteins
- PMID: 16100574
- PMCID: PMC1184040
- DOI: 10.1172/JCI25614
Schoenheimer effect explained--feedback regulation of cholesterol synthesis in mice mediated by Insig proteins
Abstract
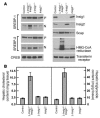
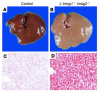
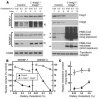
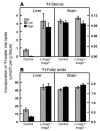
End-product feedback inhibition of cholesterol synthesis was first demonstrated in living animals by Schoenheimer 72 years ago. Current studies define Insig proteins as essential elements of this feedback system in mouse liver. In cultured cells, Insig proteins are required for sterol-mediated inhibition of the processing of sterol regulatory element-binding proteins (SREBPs) to their nuclear forms. We produced mice with germline disruption of the Insig2 gene and Cre-mediated disruption of the Insig1 gene in liver. On a chow diet, these double-knockout mice overaccumulated cholesterol and triglycerides in liver. Despite this accumulation, levels of nuclear SREBPs and mRNAs for SREBP target genes in lipogenic pathways were not reduced. Whereas cholesterol feeding reduced nuclear SREBPs and lipogenic mRNAs in wild-type mice, this feedback response was severely blunted in the double-knockout mice, and synthesis of cholesterol and fatty acids was not repressed. The amount of HMG-CoA reductase protein was elevated out of proportion to the mRNA in the double-knockout mice, apparently owing to the failure of cholesterol to accelerate degradation of the enzyme. These studies indicate that the essential elements of the regulatory pathway for lipid synthesis function in liver as they do in cultured cells.
Figures


References
-
- Schoenheimer R, Breusch F. Synthesis and destruction of cholesterol in the organism. J. Biol. Chem. 1933;103:439–448.
-
- Gould RG. Lipid metabolism and atherosclerosis. Am. J. Med. 1951;11:209–227. - PubMed
-
- Gould RG, Taylor CB, Hagerman JS, Warner I, Campbell DJ. Cholesterol metabolism. I. Effect of dietary cholesterol on the synthesis of cholesterol in dog tissue in vitro. J. Biol. Chem. 1953;201:519–523. - PubMed
-
- Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 1980;21:505–517. - PubMed
-
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor [review] Cell. 1997;89:331–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

