Highly syntenic regions in the genomes of soybean, Medicago truncatula, and Arabidopsis thaliana
- PMID: 16102170
- PMCID: PMC1201151
- DOI: 10.1186/1471-2229-5-15
Highly syntenic regions in the genomes of soybean, Medicago truncatula, and Arabidopsis thaliana
Abstract
Background: Recent genome sequencing enables mega-base scale comparisons between related genomes. Comparisons between animals, plants, fungi, and bacteria demonstrate extensive synteny tempered by rearrangements. Within the legume plant family, glimpses of synteny have also been observed. Characterizing syntenic relationships in legumes is important in transferring knowledge from model legumes to crops that are important sources of protein, fixed nitrogen, and health-promoting compounds.
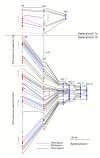
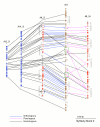
Results: We have uncovered two large soybean regions exhibiting synteny with M. truncatula and with a network of segmentally duplicated regions in Arabidopsis. In all, syntenic regions comprise over 500 predicted genes spanning 3 Mb. Up to 75% of soybean genes are colinear with M. truncatula, including one region in which 33 of 35 soybean predicted genes with database support are colinear to M. truncatula. In some regions, 60% of soybean genes share colinearity with a network of A. thaliana duplications. One region is especially interesting because this 500 kbp segment of soybean is syntenic to two paralogous regions in M. truncatula on different chromosomes. Phylogenetic analysis of individual genes within these regions demonstrates that one is orthologous to the soybean region, with which it also shows substantially denser synteny and significantly lower levels of synonymous nucleotide substitutions. The other M. truncatula region is inferred to be paralogous, presumably resulting from a duplication event preceding speciation.
Conclusion: The presence of well-defined M. truncatula segments showing orthologous and paralogous relationships with soybean allows us to explore the evolution of contiguous genomic regions in the context of ancient genome duplication and speciation events.
Figures
Similar articles
-
Fractionation of synteny in a genomic region containing tandemly duplicated genes across glycine max, Medicago truncatula, and Arabidopsis thaliana.J Hered. 2008 Jul-Aug;99(4):390-5. doi: 10.1093/jhered/esn010. Epub 2008 Mar 2. J Hered. 2008. PMID: 18316321
-
Comparative physical mapping reveals features of microsynteny between Glycine max, Medicago truncatula, and Arabidopsis thaliana.Genome. 2004 Feb;47(1):141-55. doi: 10.1139/g03-106. Genome. 2004. PMID: 15060611
-
The lipoxygenase gene family: a genomic fossil of shared polyploidy between Glycine max and Medicago truncatula.BMC Plant Biol. 2008 Dec 23;8:133. doi: 10.1186/1471-2229-8-133. BMC Plant Biol. 2008. PMID: 19105811 Free PMC article.
-
Translating Medicago truncatula genomics to crop legumes.Curr Opin Plant Biol. 2009 Apr;12(2):193-201. doi: 10.1016/j.pbi.2008.11.005. Epub 2009 Jan 21. Curr Opin Plant Biol. 2009. PMID: 19162532 Review.
-
Three sequenced legume genomes and many crop species: rich opportunities for translational genomics.Plant Physiol. 2009 Nov;151(3):970-7. doi: 10.1104/pp.109.144659. Epub 2009 Sep 16. Plant Physiol. 2009. PMID: 19759344 Free PMC article. Review. No abstract available.
Cited by
-
A BAC based physical map and genome survey of the rice false smut fungus Villosiclava virens.BMC Genomics. 2013 Dec 16;14:883. doi: 10.1186/1471-2164-14-883. BMC Genomics. 2013. PMID: 24341590 Free PMC article.
-
Codon usage vis-a-vis start and stop codon context analysis of three dicot species.J Genet. 2018 Mar;97(1):97-107. J Genet. 2018. PMID: 29666329
-
Comparative genomics to bridge Vicia faba with model and closely-related legume species: stability of QTLs for flowering and yield-related traits.Theor Appl Genet. 2012 Dec;125(8):1767-82. doi: 10.1007/s00122-012-1952-1. Epub 2012 Aug 5. Theor Appl Genet. 2012. PMID: 22864387
-
Development of gene-based molecular markers tagging low alkaloid pauper locus in white lupin (Lupinus albus L.).J Appl Genet. 2019 Nov;60(3-4):269-281. doi: 10.1007/s13353-019-00508-9. Epub 2019 Aug 13. J Appl Genet. 2019. PMID: 31410824 Free PMC article.
-
Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery.BMC Genomics. 2011 May 25;12:265. doi: 10.1186/1471-2164-12-265. BMC Genomics. 2011. PMID: 21609489 Free PMC article.
References
-
- Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, Maskeri B, Hansen NF, Schwartz MS, Weber RJ, Kent WJ, Karolchik D, Bruen TC, Bevan R, Cutler DJ, Schwartz S, Elnitski L, Idol JR, Prasad AB, Lee-Lin SQ, Maduro VV, Summers TJ, Portnoy ME, Dietrich NL, Akhter N, Ayele K, Benjamin B, Cariaga K, Brinkley CP, Brooks SY, Granite S, Guan X, Gupta J, Haghighi P, Ho SL, Huang MC, Karlins E, Laric PL, Legaspi R, Lim MJ, Maduro QL, Masiello CA, Mastrian SD, McCloskey JC, Pearson R, Stantripop S, Tiongson EE, Tran JT, Tsurgeon C, Vogt JL, Walker MA, Wetherby KD, Wiggins LS, Young AC, Zhang LH, Osoegawa K, Zhu B, Zhao B, Shu CL, De Jong PJ, Lawrence CE, Smit AF, Chakravarti A, Haussler D, Green P, Miller W, Green ED. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

