Human coronavirus NL63, France
- PMID: 16102311
- PMCID: PMC3320486
- DOI: 10.3201/eid1108.050110
Human coronavirus NL63, France
Abstract
The human coronavirus NL63 (HCoV-NL63) was first identified in The Netherlands, and its circulation in France has not been investigated. We studied HCoV-NL63 infection in hospitalized children diagnosed with respiratory tract infections. From November 2002 to April 2003, we evaluated 300 respiratory specimens for HCoV-NL63. Of the 300 samples, 28 (9.3%) were positive for HCoV-NL63. The highest prevalence was found in February (18%). The main symptoms were fever (61%), rhinitis (39%), bronchiolitis (39%), digestive problems (33%), otitis (28%), pharyngitis (22%), and conjunctivitis (17%). A fragment of the spike protein gene was sequenced to determine the variety of circulating HCoV-NL63. Phylogenetic analysis indicated that strains with different genetic markers cocirculate in France.
Figures

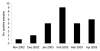

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
