The biology of human lymphoid malignancies revealed by gene expression profiling
- PMID: 16102574
- PMCID: PMC1351148
- DOI: 10.1016/S0065-2776(05)87005-1
The biology of human lymphoid malignancies revealed by gene expression profiling
Abstract
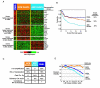
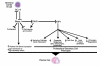
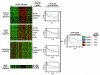
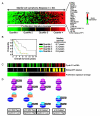
Gene expression profiling provides a quantitative molecular framework for the study of human lymphomas. This genomic technology has revealed that existing diagnostic categories are comprised of multiple molecularly and clinically distinct diseases. Diffuse large B-cell lymphoma (DLBCL), for example, consists of three gene expression subgroups, termed germinal center B-cell-like (GCB) DLBCL, activated B-cell-like (ABC) DLBCL, and primary mediastinal B-cell lymphoma (PMBL). These DLBCL subgroups arise from different stages of normal B-cell differentiation, utilize distinct oncogenic mechanisms, and differ in their ability to be cured by chemotherapy. Key regulatory factors and their target genes are differentially expressed among these subgroups, including BCL-6, Blimp-1, and XBP1. ABC DLBCL and PMBL depend upon constitutive activation of the NF-kappaB pathway for their survival but GCB DLBCL does not, demonstrating that this pathway is a potential therapeutic target for certain DLBCL subgroups. In DLBCL, mantle cell lymphoma, and follicular lymphoma, gene expression profiling has also been used to create gene expression-based models of survival, which have identified the biological characteristics of the tumors that influence their clinical behavior. In mantle cell lymphoma, the length of survival following diagnosis is primarily influenced by the tumor proliferation rate, which can be quantitatively measured by a proliferation gene expression "signature." Based on this accurate measure, the proliferation rate can now be viewed as an integration of several oncogenic lesions that each increase progression from the G1 to the S phase of the cell cycle. In DLBCL and follicular lymphoma, gene expression profiling has revealed that the molecular characteristics of non-malignant tumor-infiltrating immune cells have a major influence on the length of survival. The implications of these insights for the diagnosis and treatment of non-Hodgkin lymphomas are discussed.
Figures
References
-
- Addis BJ, Isaacson PG. Histopathology. 1986;10:379. - PubMed
-
- Albagli O, Dhordain P, Bernardin F, Quief S, Kerkaert JP, Leprince D. Biochem Biophys Res Commun. 1996;220:911. - PubMed
-
- Alizadeh A, Eisen M, Davis RE, Ma C, Sabet H, Tran T, Powell J, Yang L, Marti G, Moore T, Hudson J, Chan WC, Greiner T, Weisenburger D, Armitage JO, Lossos I, Levy R, Botstein D, Brown PO, Staudt LM. Cold Spring Harbor Symp Quant Biol. 1999;64:71. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Nature. 2000;403:503. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

