Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection
- PMID: 16102740
- PMCID: PMC7124242
- DOI: 10.1016/j.cccn.2005.06.023
Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection
Abstract
Background: As we enter the post-genome sequencing era and begin to sift through the enormous amount of genetic information now available, the need for technologies that allow rapid, cost-effective, high-throughput detection of specific nucleic acid sequences becomes apparent. Multiplexing technologies, which allow for simultaneous detection of multiple nucleic acid sequences in a single reaction, can greatly reduce the time, cost and labor associated with single reaction detection technologies.
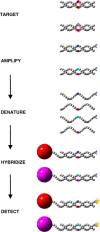
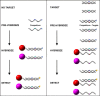
Methods: The Luminex xMAP system is a multiplexed microsphere-based suspension array platform capable of analyzing and reporting up to 100 different reactions in a single reaction vessel. This technology provides a new platform for high-throughput nucleic acid detection and is being utilized with increasing frequency. Here we review specific applications of xMAP technology for nucleic acid detection in the areas of single nucleotide polymorphism (SNP) genotyping, genetic disease screening, gene expression profiling, HLA DNA typing and microbial detection.
Conclusions: These studies demonstrate the speed, efficiency and utility of xMAP technology for simultaneous, rapid, sensitive and specific nucleic acid detection, and its capability to meet the current and future requirements of the molecular laboratory for high-throughput nucleic acid detection.
Figures


References
-
- Collins F.S., Guyer M.S., Chakravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
-
- Collins F.S., McKusick V.A. Implications of the Human Genome Project for medical science. JAMA. 2001;285:540–544. - PubMed
-
- Cotton R.G. Methods in clinical molecular genetics. Eur J Pediatr. 2000;159(Suppl. 3):179–182. - PubMed
-
- Ugozolli L.A. Multiplex assays with fluorescent microbead readout: a powerful tool for mutation detection. Clin Chem. 2004;50:1963–1965. - PubMed
-
- Fulton R.J., McDade R.L., Smith P.L., Kienker L.J., Kettman J.R., Jr. Advanced multiplexed analysis with the FlowMetrix™ system. Clin Chem. 1997;43:1749–1756. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

