Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice
- PMID: 16103145
- PMCID: PMC1193591
- DOI: 10.1128/JVI.79.17.10931-10943.2005
Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice
Abstract
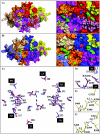
Two strains of the parvovirus minute virus of mice (MVM), the immunosuppressive (MVMi) and the prototype (MVMp) strains, display disparate in vitro tropism and in vivo pathogenicity. We report the crystal structures of MVMp virus-like particles (MVMp(b)) and native wild-type (wt) empty capsids (MVMp(e)), determined and refined to 3.25 and 3.75 A resolution, respectively, and their comparison to the structure of MVMi, also refined to 3.5 A resolution in this study. A comparison of the MVMp(b) and MVMp(e) capsids showed their structures to be the same, providing structural verification that some heterologously expressed parvovirus capsids are indistinguishable from wt capsids produced in host cells. The structures of MVMi and MVMp capsids were almost identical, but local surface conformational differences clustered from symmetry-related capsid proteins at three specific domains: (i) the icosahedral fivefold axis, (ii) the "shoulder" of the protrusion at the icosahedral threefold axis, and (iii) the area surrounding the depression at the icosahedral twofold axis. The latter two domains contain important determinants of MVM in vitro tropism (residues 317 and 321) and forward mutation residues (residues 399, 460, 553, and 558) conferring fibrotropism on MVMi. Furthermore, these structural differences between the MVM strains colocalize with tropism and pathogenicity determinants mapped for other autonomous parvovirus capsids, highlighting the importance of common parvovirus capsid regions in the control of virus-host interactions.
Figures



Similar articles
-
Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles.Virology. 2000 Feb 15;267(2):299-309. doi: 10.1006/viro.1999.0123. Virology. 2000. PMID: 10662625
-
Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation.J Biol Chem. 2006 Sep 1;281(35):25670-7. doi: 10.1074/jbc.M604421200. Epub 2006 Jul 5. J Biol Chem. 2006. PMID: 16822863
-
Functional implications of the structure of the murine parvovirus, minute virus of mice.Structure. 1998 Nov 15;6(11):1369-81. doi: 10.1016/s0969-2126(98)00137-3. Structure. 1998. PMID: 9817841
-
Parvoviral host range and cell entry mechanisms.Adv Virus Res. 2007;70:183-232. doi: 10.1016/S0065-3527(07)70005-2. Adv Virus Res. 2007. PMID: 17765706 Review.
-
Twenty-Five Years of Structural Parvovirology.Viruses. 2019 Apr 20;11(4):362. doi: 10.3390/v11040362. Viruses. 2019. PMID: 31010002 Free PMC article. Review.
Cited by
-
Structural Insights into Human Bocaparvoviruses.J Virol. 2017 May 12;91(11):e00261-17. doi: 10.1128/JVI.00261-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331084 Free PMC article.
-
Retargeting of rat parvovirus H-1PV to cancer cells through genetic engineering of the viral capsid.J Virol. 2012 Apr;86(7):3452-65. doi: 10.1128/JVI.06208-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258256 Free PMC article.
-
Resolving structure and mechanical properties at the nanoscale of viruses with frequency modulation atomic force microscopy.PLoS One. 2012;7(1):e30204. doi: 10.1371/journal.pone.0030204. Epub 2012 Jan 25. PLoS One. 2012. PMID: 22295076 Free PMC article.
-
Minute virus of mice, a parvovirus, in complex with the Fab fragment of a neutralizing monoclonal antibody.J Virol. 2007 Sep;81(18):9851-8. doi: 10.1128/JVI.00775-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626084 Free PMC article.
-
Protoparvovirus Cell Entry.Viruses. 2017 Oct 26;9(11):313. doi: 10.3390/v9110313. Viruses. 2017. PMID: 29072600 Free PMC article. Review.
References
-
- Agbandje, M., S. Kajigaya, R. McKenna, N. S. Young, and M. G. Rossmann. 1994. The structure of human parvovirus B19 at 8 Å resolution. Virology 203:106-115. - PubMed
-
- Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and P. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. - PubMed
-
- Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. - PubMed
-
- Agbandje-McKenna, M., C. R. Parrish, and M. G. Rossmann. 1995. The structure of parvoviruses. Semin. Virol. 6:299-309.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

