Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes
- PMID: 16103176
- PMCID: PMC1193597
- DOI: 10.1128/JVI.79.17.11239-11246.2005
Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes
Abstract
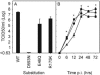
Viruses can exploit a variety of strategies to evade immune surveillance by cytotoxic T lymphocytes (CTL), including the acquisition of mutations in CTL epitopes. Also for influenza A viruses a number of amino acid substitutions in the nucleoprotein (NP) have been associated with escape from CTL. However, other previously identified influenza A virus CTL epitopes are highly conserved, including the immunodominant HLA-A*0201-restricted epitope from the matrix protein, M1(58-66). We hypothesized that functional constraints were responsible for the conserved nature of influenza A virus CTL epitopes, limiting escape from CTL. To assess the impact of amino acid substitutions in conserved epitopes on viral fitness and recognition by specific CTL, we performed a mutational analysis of CTL epitopes. Both alanine replacements and more conservative substitutions were introduced at various positions of different influenza A virus CTL epitopes. Alanine replacements for each of the nine amino acids of the M1(58-66) epitope were tolerated to various extents, except for the anchor residue at the second position. Substitution of anchor residues in other influenza A virus CTL epitopes also affected viral fitness. Viable mutant viruses were used in CTL recognition experiments. The results are discussed in the light of the possibility of influenza viruses to escape from specific CTL. It was speculated that functional constraints limit variation in certain epitopes, especially at anchor residues, explaining the conserved nature of these epitopes.
Figures
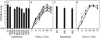



References
-
- Bednarek, M. A., S. Y. Sauma, M. C. Gammon, G. Porter, S. Tamhankar, A. R. Williamson, and H. J. Zweerink. 1991. The minimum peptide epitope from the influenza virus matrix protein. Extra and intracellular loading of HLA-A2. J. Immunol. 147:4047-4053. - PubMed
-
- Berkhoff, E. G., A. C. M. Boon, N. J. Nieuwkoop, R. A. Fouchier, K. Sintnicolaas, A. D. M. E. Osterhaus, and G. F. Rimmelzwaan. 2004. A mutation in the HLA-B*2705-restricted NP383-391 epitope affects the human influenza A virus-specific cytotoxic T-lymphocyte response in vitro. J. Virol. 78:5216-5222. - PMC - PubMed
-
- Boon, A. C., G. de Mutsert, Y. M. Graus, R. A. Fouchier, K. Sintnicolaas, A. D. M. E. Osterhaus, and G. F. Rimmelzwaan. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76:582-590. - PMC - PubMed
-
- Boon, A. C. M., G. de Mutsert, Y. M. F. Graus, R. A. M. Fouchier, K. Sintnicolaas, A. D. M. E. Osterhaus, and G. F. Rimmelzwaan. 2002. Sequence variation in a newly identified HLA-B35-restricted epitope in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. J. Virol. 76:2567-2572. - PMC - PubMed
-
- Boon, A. C. M., G. de Mutsert, D. van Baarle, D. J. Smith, A. S. Lapedes, R. A. Fouchier, K. Sintnicolaas, A. D. Osterhaus, and G. F. Rimmelzwaan. 2004. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J. Immunol. 172:2453-2460. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

