The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation
- PMID: 16103198
- PMCID: PMC1193639
- DOI: 10.1128/JVI.79.17.11476-11486.2005
The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation
Abstract
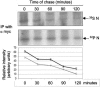
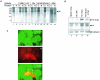
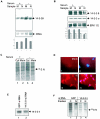
The severe acute respiratory syndrome coronavirus(SARS-CoV) nucleocapsid (N) protein is one of the four structural proteins of the virus and is predicted to be a 46-kDa phosphoprotein. Our in silico analysis predicted N to be heavily phosphorylated at multiple residues. Experimentally, we have shown in this report that the N protein of the SARS-CoV gets serine-phosphorylated by multiple kinases, in both the cytoplasm and the nucleus. The phosphoprotein is stable and localizes in the cytoplasm and coprecipitates with the membrane fraction. Also, using specific inhibitors of phosphorylation and an in vitro phosphorylation assay, we show that the nucleocapsid protein is a substrate of cyclin-dependent kinase (CDK), glycogen synthase kinase, mitogen-activated protein kinase, and casein kinase II. Further, we show that the phosphorylated protein is translocated to the cytoplasm by binding to 14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein). 14-3-3 proteins are a family of highly conserved, ubiquitously expressed eukaryotic proteins that function primarily as adapters that modulate interactions between components of various cellular signaling and cell cycle regulatory pathways through phosphorylation-dependent protein-protein interactions. Coincidentally, the N protein was also found to downregulate the expression of the theta isoform of 14-3-3 (14-3-3theta), leading to the accumulation of phosphorylated N protein in the nucleus, in the absence of growth factors. Using short interfering RNA specific to 14-3-3theta we have inhibited its expression to show accumulation of phosphorylated N protein in the nucleus. Thus, the data presented here provide a possible mechanism for phosphorylation-dependent nucleocytoplasmic shuttling of the N protein. This 14-3-3-mediated transport of the phosphorylated N protein and its possible implications in interfering with the cellular machinery are discussed.
Figures


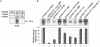

References
-
- Buttner, J. 1977. Evaluation of the diagnostic value of laboratory investigations. J. Clin. Chem. Clin. Biochem. 15:1-12. - PubMed
-
- Centers for Disease Control SARS Investigative Team and A. T. Fleischauer. 2003. Outbreak of severe acute respiratory syndrome -worldwide. Morb. Mortal. Wkly. Rep. 52:226-228. - PubMed
-
- Chijiwa, T., A. Mishima, M. Hagiwara, M. Sano, K. Hayashi, T. Inoue, K. Naito, T. Toshioka, and H. Hidaka. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265:5267-5272. - PubMed
-
- Donnelly, C. A., A. C. Ghani, G. M. Leung, A. J. Hedley, C. Fraser, S. Riley, L. J. Abu-Raddad, L. M. Ho, T. Q. Thach, P. Chau, K. P. Chan, T. H. Lam, L. Y. Tse, T. Tsang, S. H. Liu, J. H. Kong, E. M. Lau, N. M. Ferguson, and R. M. Anderson. 2003. Epidemiological determinants of spread of causative agent of severe acute respiratory syndrome in Hong Kong. Lancet 361:1761-1766. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

