Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications
- PMID: 16103216
- PMCID: PMC1186188
- DOI: 10.1101/gad.1333905
Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications
Abstract
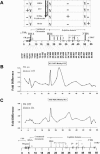
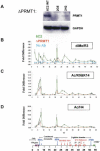
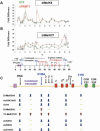
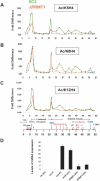
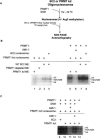
PRMT1 is a histone methyltransferase that methylates Arg3 on histone H4. When we used siRNA to knock down PRMT1 in an erythroid cell line, it resulted in nearly complete loss of H4 Arg3 methylation across the chicken beta-globin domain, which we use as a model system for studying the relationship of gene activity to histone modification. We observed furthermore a domain-wide loss of histone acetylation on both histones H3 and H4, as well as an increase in H3 Lys9 and Lys27 methylation, both marks associated with inactive chromatin. To determine whether the effect on acetylation was directly related to the loss of H4 Arg3 methylation, we performed an in vitro acetylation reaction on chromatin isolated from PRMT1-depleted cells. We found that nucleosomes purified from these cells, and depleted in methylation at Arg3, are readily acetylated by nuclear extracts from the same cells, if and only if the nucleosomes are incubated with PRMT1 beforehand. Thus, methylation of histones by PRMT1 was sufficient to permit subsequent acetylation. Consistent with earlier reports of experiments in vitro, H4 Arg3 methylation by PRMT1 appears to be essential in vivo for the establishment or maintenance of a wide range of "active" chromatin modifications.
Figures
References
-
- Agalioti T., Chen, G., and Thanos, D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392. - PubMed
-
- An W., Kim, J., and Roeder, R.G. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748. - PubMed
-
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
-
- Bell A.C., West, A.G., and Felsenfeld, G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
