Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila
- PMID: 16103923
- PMCID: PMC1183528
- DOI: 10.1371/journal.pgen.0010009
Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila
Abstract
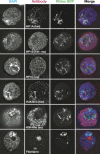
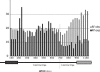
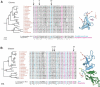
Heterochromatin comprises a significant component of many eukaryotic genomes. In comparison to euchromatin, heterochromatin is gene poor, transposon rich, and late replicating. It serves many important biological roles, from gene silencing to accurate chromosome segregation, yet little is known about the evolutionary constraints that shape heterochromatin. A complementary approach to the traditional one of directly studying heterochromatic DNA sequence is to study the evolution of proteins that bind and define heterochromatin. One of the best markers for heterochromatin is the heterochromatin protein 1 (HP1), which is an essential, nonhistone chromosomal protein. Here we investigate the molecular evolution of five HP1 paralogs present in Drosophila melanogaster. Three of these paralogs have ubiquitous expression patterns in adult Drosophila tissues, whereas HP1D/rhino and HP1E are expressed predominantly in ovaries and testes respectively. The HP1 paralogs also have distinct localization preferences in Drosophila cells. Thus, Rhino localizes to the heterochromatic compartment in Drosophila tissue culture cells, but in a pattern distinct from HP1A and lysine-9 dimethylated H3. Using molecular evolution and population genetic analyses, we find that rhino has been subject to positive selection in all three domains of the protein: the N-terminal chromo domain, the C-terminal chromo-shadow domain, and the hinge region that connects these two modules. Maximum likelihood analysis of rhino sequences from 20 species of Drosophila reveals that a small number of residues of the chromo and shadow domains have been subject to repeated positive selection. The rapid and positive selection of rhino is highly unusual for a gene encoding a chromosomal protein and suggests that rhino is involved in a genetic conflict that affects the germline, belying the notion that heterochromatin is simply a passive recipient of "junk DNA" in eukaryotic genomes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


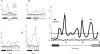

Similar articles
-
The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins.Mol Cell Biol. 2001 Apr;21(7):2555-69. doi: 10.1128/MCB.21.7.2555-2569.2001. Mol Cell Biol. 2001. PMID: 11259603 Free PMC article.
-
Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family.PLoS Genet. 2012;8(6):e1002729. doi: 10.1371/journal.pgen.1002729. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737079 Free PMC article.
-
The Drosophila ZAD zinc finger protein Kipferl guides Rhino to piRNA clusters.Elife. 2022 Oct 4;11:e80067. doi: 10.7554/eLife.80067. Elife. 2022. PMID: 36193674 Free PMC article.
-
Multiple roles for heterochromatin protein 1 genes in Drosophila.Annu Rev Genet. 2009;43:467-92. doi: 10.1146/annurev-genet-102108-134802. Annu Rev Genet. 2009. PMID: 19919324 Review.
-
Does heterochromatin protein 1 always follow code?Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16462-9. doi: 10.1073/pnas.162371699. Epub 2002 Jul 31. Proc Natl Acad Sci U S A. 2002. PMID: 12151603 Free PMC article. Review.
Cited by
-
Structural insights into Rhino-mediated germline piRNA cluster formation.Cell Res. 2015 Apr;25(4):525-8. doi: 10.1038/cr.2015.10. Epub 2015 Jan 23. Cell Res. 2015. PMID: 25613572 Free PMC article. No abstract available.
-
Functions of HP1 proteins in transcriptional regulation.Epigenetics Chromatin. 2022 May 7;15(1):14. doi: 10.1186/s13072-022-00453-8. Epigenetics Chromatin. 2022. PMID: 35526078 Free PMC article. Review.
-
Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei.Plant Cell. 2006 Jan;18(1):133-45. doi: 10.1105/tpc.105.036855. Epub 2005 Dec 16. Plant Cell. 2006. PMID: 16361394 Free PMC article.
-
Multiple functions for Drosophila Mcm10 suggested through analysis of two Mcm10 mutant alleles.Genetics. 2010 Aug;185(4):1151-65. doi: 10.1534/genetics.110.117234. Epub 2010 May 24. Genetics. 2010. PMID: 20498296 Free PMC article.
-
Adaptive Evolution Targets a piRNA Precursor Transcription Network.Cell Rep. 2020 Feb 25;30(8):2672-2685.e5. doi: 10.1016/j.celrep.2020.01.109. Cell Rep. 2020. PMID: 32101744 Free PMC article.
References
-
- Hodgetts R. Eukaryotic gene regulation by targeted chromatin re-modeling at dispersed, middle-repetitive sequence elements. Curr Opin Genet Dev. 2004;14:680–685. - PubMed
-
- Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. - PubMed
-
- Malik HS, Henikoff S. Conflict begets complexity: The evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. - PubMed
-
- Choo KH. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–177. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

