p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest
- PMID: 16107691
- PMCID: PMC1190302
- DOI: 10.1128/MCB.25.17.7423-7431.2005
p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest
Abstract
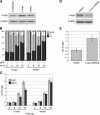
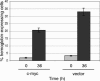
The ability of p53 to promote apoptosis and cell cycle arrest is believed to be important for its tumor suppression function. Besides activating the expression of cell cycle arrest and proapoptotic genes, p53 also represses a number of genes. Previous studies have shown an association between p53 activation and down-regulation of c-myc expression. However, the mechanism and physiological significance of p53-mediated c-myc repression remain unclear. Here, we show that c-myc is repressed in a p53-dependent manner in various mouse and human cell lines and mouse tissues. Furthermore, c-myc repression is not dependent on the expression of p21(WAF1). Abrogating the repression of c-myc by ectopic c-myc expression interferes with the ability of p53 to induce G(1) cell cycle arrest and differentiation but enhances the ability of p53 to promote apoptosis. We propose that p53-dependent cell cycle arrest is dependent not only on the transactivation of cell cycle arrest genes but also on the transrepression of c-myc. Chromatin immunoprecipitation assays indicate that p53 is bound to the c-myc promoter in vivo. We report that trichostatin A, an inhibitor of histone deacetylases, abrogates the ability of p53 to repress c-myc transcription. We also show that p53-mediated transcriptional repression of c-myc is accompanied by a decrease in the level of acetylated histone H4 at the c-myc promoter and by recruitment of the corepressor mSin3a. These data suggest that p53 represses c-myc transcription through a mechanism that involves histone deacetylation.
Figures





References
-
- Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. - PubMed
-
- Coppola, J. A., and M. D. Cole. 1986. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature 320:760-763. - PubMed
-
- D'Souza, S., H. Xin, S. Walter, and D. Choubey. 2001. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J. Biol. Chem. 276:298-305. - PubMed
-
- Eberhardy, S. R., C. A. D'Cunha, and P. J. Farnham. 2000. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J. Biol. Chem. 275:33798-33805. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
