The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection
- PMID: 16107718
- PMCID: PMC1190295
- DOI: 10.1128/MCB.25.17.7725-7733.2005
The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection
Abstract
Pot1 (protection of telomeres 1) is a single-stranded telomere binding protein that is essential for chromosome end protection and telomere length homeostasis. Arabidopsis encodes two Pot1-like proteins, dubbed AtPot1 and AtPot2. Here we show that telomeres in transgenic plants expressing a truncated AtPot1 allele lacking the N-terminal oligonucleotide/oligosaccharide binding fold (P1DeltaN) are 1 to 1.5 kb shorter than in the wild type, suggesting that AtPot1 contributes to the positive regulation of telomere length control. In contrast, telomere length is unperturbed in plants expressing the analogous region of AtPot2. A strikingly different phenotype is observed in plants overexpressing the AtPot2 N terminus (P2DeltaC) but not the corresponding region in AtPot1. Although bulk telomeres in P2DeltaC mutants are 1 to 2 kb shorter than in the wild type, these plants resemble late-generation telomerase-deficient mutants with severe growth defects, sterility, and massive genome instability, including bridged chromosomes and aneuploidy. The genome instability associated with P2DeltaC mutants implies that AtPot2 contributes to chromosome end protection. Thus, Arabidopsis has evolved two Pot genes that function differently in telomere biology. These findings provide unanticipated information about the evolution of single-stranded telomere binding proteins.
Figures

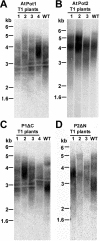
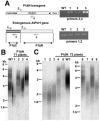
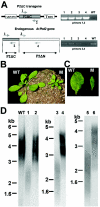

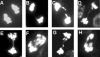
Similar articles
-
Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance.EMBO J. 2007 Aug 8;26(15):3653-61. doi: 10.1038/sj.emboj.7601792. Epub 2007 Jul 12. EMBO J. 2007. PMID: 17627276 Free PMC article.
-
Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis.Plant J. 2007 Feb;49(3):442-51. doi: 10.1111/j.1365-313X.2006.02974.x. Epub 2007 Jan 1. Plant J. 2007. PMID: 17217467
-
Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability.Mol Cell Biol. 2005 Jul;25(13):5567-78. doi: 10.1128/MCB.25.13.5567-5578.2005. Mol Cell Biol. 2005. PMID: 15964812 Free PMC article.
-
POT1 mutations cause differential effects on telomere length leading to opposing disease phenotypes.J Cell Physiol. 2023 Jun;238(6):1237-1255. doi: 10.1002/jcp.31034. Epub 2023 May 14. J Cell Physiol. 2023. PMID: 37183325 Review.
-
Cytogenetics for the study of telomere function in plants.Cytogenet Genome Res. 2008;122(3-4):374-9. doi: 10.1159/000167825. Epub 2009 Jan 30. Cytogenet Genome Res. 2008. PMID: 19188708 Review.
Cited by
-
POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination.EMBO J. 2006 Nov 1;25(21):5180-90. doi: 10.1038/sj.emboj.7601294. Epub 2006 Oct 19. EMBO J. 2006. PMID: 17053789 Free PMC article.
-
Functional dissection of human and mouse POT1 proteins.Mol Cell Biol. 2009 Jan;29(2):471-82. doi: 10.1128/MCB.01352-08. Epub 2008 Oct 27. Mol Cell Biol. 2009. PMID: 18955498 Free PMC article.
-
POT1a and components of CST engage telomerase and regulate its activity in Arabidopsis.PLoS Genet. 2014 Oct 16;10(10):e1004738. doi: 10.1371/journal.pgen.1004738. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25329641 Free PMC article.
-
Telomere- and Telomerase-Associated Proteins and Their Functions in the Plant Cell.Front Plant Sci. 2016 Jun 28;7:851. doi: 10.3389/fpls.2016.00851. eCollection 2016. Front Plant Sci. 2016. PMID: 27446102 Free PMC article. Review.
-
The epigenetic regulation of centromeres and telomeres in plants and animals.Comp Cytogenet. 2020 Jul 7;14(2):265-311. doi: 10.3897/CompCytogen.v14i2.51895. eCollection 2020. Comp Cytogenet. 2020. PMID: 32733650 Free PMC article. Review.
References
-
- Armstrong, S. J., F. C. Franklin, and G. H. Jones. 2001. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114:4207-4217. - PubMed
-
- Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. - PubMed
-
- Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
