CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin
- PMID: 16107875
- PMCID: PMC1224683
- DOI: 10.1038/sj.emboj.7600793
CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin
Abstract
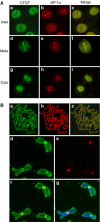
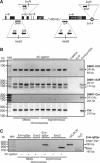
Most of the transcription factors, RNA polymerases and enhancer binding factors are absent from condensed mitotic chromosomes. In contrast, epigenetic marks of active and inactive genes somehow survive mitosis, since the activity status from one cell generation to the next is maintained. For the zinc-finger protein CTCF, a role in interpreting and propagating epigenetic states and in separating expression domains has been documented. To test whether such a domain structure is preserved during mitosis, we examined whether CTCF is bound to mitotic chromatin. Here we show that in contrast to other zinc-finger proteins, CTCF indeed is bound to mitotic chromosomes. Mitotic binding is mediated by a portion of the zinc-finger DNA binding domain and involves sequence specific binding to target sites. Furthermore, the chromatin loop organized by the CTCF-bound, differentially methylated region at the Igf2/H19 locus can be detected in mitosis. In contrast, the enhancer/promoter loop of the same locus is lost in mitosis. This may provide a novel form of epigenetic memory during cell division.
Figures



References
-
- Adams RR, Carmena M, Earnshaw WC (2001) Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol 11: 49–54 - PubMed
-
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 - PubMed
-
- Berube NG, Smeenk CA, Picketts DJ (2000) Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet 9: 539–547 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

