Inactivation of the Huntington's disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo
- PMID: 16109169
- PMCID: PMC1208873
- DOI: 10.1186/1471-213X-5-17
Inactivation of the Huntington's disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo
Abstract
Background: Huntingtin, the HD gene encoded protein mutated by polyglutamine expansion in Huntington's disease, is required in extraembryonic tissues for proper gastrulation, implicating its activities in nutrition or patterning of the developing embryo. To test these possibilities, we have used whole mount in situ hybridization to examine embryonic patterning and morphogenesis in homozygous Hdh(ex4/5) huntingtin deficient embryos.
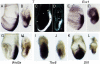
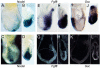
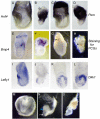
Results: In the absence of huntingtin, expression of nutritive genes appears normal but E7.0-7.5 embryos exhibit a unique combination of patterning defects. Notable are a shortened primitive streak, absence of a proper node and diminished production of anterior streak derivatives. Reduced Wnt3a, Tbx6 and Dll1 expression signify decreased paraxial mesoderm and reduced Otx2 expression and lack of headfolds denote a failure of head development. In addition, genes initially broadly expressed are not properly restricted to the posterior, as evidenced by the ectopic expression of Nodal, Fgf8 and Gsc in the epiblast and T (Brachyury) and Evx1 in proximal mesoderm derivatives. Despite impaired posterior restriction and anterior streak deficits, overall anterior/posterior polarity is established. A single primitive streak forms and marker expression shows that the anterior epiblast and anterior visceral endoderm (AVE) are specified.
Conclusion: Huntingtin is essential in the early patterning of the embryo for formation of the anterior region of the primitive streak, and for down-regulation of a subset of dynamic growth and transcription factor genes. These findings provide fundamental starting points for identifying the novel cellular and molecular activities of huntingtin in the extraembryonic tissues that govern normal anterior streak development. This knowledge may prove to be important for understanding the mechanism by which the dominant polyglutamine expansion in huntingtin determines the loss of neurons in Huntington's disease.
Figures

References
-
- MacDonald ME. Huntingtin: alive and well and working in middle management. Sci STKE. 2003;2003:pe48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

