A phylogeny of bacterial RNA nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases
- PMID: 16109934
- PMCID: PMC1196141
- DOI: 10.1128/JB.187.17.5927-5936.2005
A phylogeny of bacterial RNA nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases
Abstract
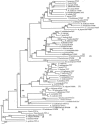
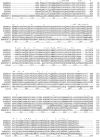
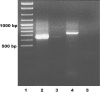
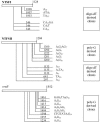
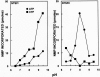
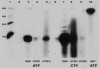
We have analyzed the distribution of RNA nucleotidyltransferases from the family that includes poly(A) polymerases (PAP) and tRNA nucleotidyltransferases (TNT) in 43 bacterial species. Genes of several bacterial species encode only one member of the nucleotidyltransferase superfamily (NTSF), and if that protein functions as a TNT, those organisms may not contain a poly(A) polymerase I like that of Escherichia coli. The genomes of several of the species examined encode more than one member of the nucleotidyltransferase superfamily. The function of some of those proteins is known, but in most cases no biochemical activity has been assigned to the NTSF. The NTSF protein sequences were used to construct an unrooted phylogenetic tree. To learn more about the function of the NTSFs in species whose genomes encode more than one, we have examined Bacillus halodurans. We have demonstrated that B. halodurans adds poly(A) tails to the 3' ends of RNAs in vivo. We have shown that the genes for both of the NTSFs encoded by the B. halodurans genome are transcribed in vivo. We have cloned, overexpressed, and purified the two NTSFs and have shown that neither functions as poly(A) polymerase in vitro. Rather, the two proteins function as tRNA nucleotidyltransferases, and our data suggest that, like some of the deep branching bacterial species previously studied by others, B. halodurans possesses separate CC- and A-adding tRNA nucleotidyltransferases. These observations raise the interesting question of the identity of the enzyme responsible for RNA polyadenylation in Bacillus.
Figures

References
-
- Bocchetta, M., S. Gribaldo, A. Sanangelantoni, and P. Cammarano. 2000. Phylogenetic depth of the bacterial genera Aquifex and Thermotoga inferred from analysis of ribosomal protein, elongation factor and RNA polymerase subunit sequences. J. Mol. Evol. 50:366-380. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Bralley, P., and G. H. Jones. 2002. cDNA cloning confirms the polyadenylation of RNA decay intermediates in Streptomyces coelicolor. Microbiology 148:1421-1425. - PubMed
-
- Bralley, P., and G. H. Jones. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149:2173-2182. - PubMed
-
- Bralley, P., and G. H. Jones. 2001. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:1155-1164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

