Exploring Lactobacillus plantarum genome diversity by using microarrays
- PMID: 16109953
- PMCID: PMC1196139
- DOI: 10.1128/JB.187.17.6119-6127.2005
Exploring Lactobacillus plantarum genome diversity by using microarrays
Abstract
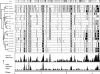
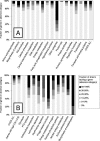
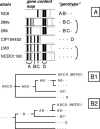
Lactobacillus plantarum is a versatile and flexible species that is encountered in a variety of niches and can utilize a broad range of fermentable carbon sources. To assess if this versatility is linked to a variable gene pool, microarrays containing a subset of small genomic fragments of L. plantarum strain WCFS1 were used to perform stringent genotyping of 20 strains of L. plantarum from various sources. The gene categories with the most genes conserved in all strains were those involved in biosynthesis or degradation of structural compounds like proteins, lipids, and DNA. Conversely, genes involved in sugar transport and catabolism were highly variable between strains. Moreover, besides the obvious regions of variance, like prophages, other regions varied between the strains, including regions encoding plantaricin biosynthesis, nonribosomal peptide biosynthesis, and exopolysaccharide biosynthesis. In many cases, these variable regions colocalized with regions of unusual base composition. Two large regions of flexibility were identified between 2.70 and 2.85 and 3.10 and 3.29 Mb of the WCFS1 chromosome, the latter being close to the origin of replication. The majority of genes encoded in these variable regions are involved in sugar metabolism. This functional overrepresentation and the unusual base composition of these regions led to the hypothesis that they represented lifestyle adaptation regions in L. plantarum. The present study consolidates this hypothesis by showing that there is a high degree of gene content variation among L. plantarum strains in genes located in these regions of the WCFS1 genome. Interestingly, based on our genotyping data L. plantarum strains clustered into two clearly distinguishable groups, which coincided with an earlier proposed subdivision of this species based on conventional methods.
Figures
References
-
- Ahrne, S., S. Nobaek, B. Jeppsson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88-94. - PubMed
-
- Aukrust, T., and H. Blom. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253-261.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

