Characterization of the genome composition of Bartonella koehlerae by microarray comparative genomic hybridization profiling
- PMID: 16109957
- PMCID: PMC1196136
- DOI: 10.1128/JB.187.17.6155-6165.2005
Characterization of the genome composition of Bartonella koehlerae by microarray comparative genomic hybridization profiling
Abstract
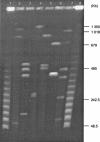
Bartonella henselae is present in a wide range of wild and domestic feline hosts and causes cat-scratch disease and bacillary angiomatosis in humans. We have estimated here the gene content of Bartonella koehlerae, a novel species isolated from cats that was recently identified as an agent of human endocarditis. The investigation was accomplished by comparative genomic hybridization (CGH) to a microarray constructed from the sequenced 1.93-Mb genome of B. henselae. Control hybridizations of labeled DNA from the human pathogen Bartonella quintana with a reduced genome of 1.58 Mb were performed to evaluate the accuracy of the array for genes with known levels of sequence divergence. Genome size estimates of B. koehlerae by pulsed-field gel electrophoresis matched that calculated by the CGH, indicating a genome of 1.7 to 1.8 Mb with few unique genes. As in B. quintana, sequences in the prophage and the genomic islands were reported absent in B. koehlerae. In addition, sequence variability was recorded in the chromosome II-like region, where B. koehlerae showed an intermediate retention pattern of both coding and noncoding sequences. Although most of the genes missing in B. koehlerae are also absent from B. quintana, its phylogenetic placement near B. henselae suggests independent deletion events, indicating that host specificity is not solely attributed to genes in the genomic islands. Rather, the results underscore the instability of the genomic islands even within bacterial populations adapted to the same host-vector system, as in the case of B. henselae and B. koehlerae.
Figures



Similar articles
-
Genome dynamics of Bartonella grahamii in micro-populations of woodland rodents.BMC Genomics. 2010 Mar 4;11:152. doi: 10.1186/1471-2164-11-152. BMC Genomics. 2010. PMID: 20202191 Free PMC article.
-
Genome rearrangements, deletions, and amplifications in the natural population of Bartonella henselae.J Bacteriol. 2006 Nov;188(21):7426-39. doi: 10.1128/JB.00472-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936024 Free PMC article.
-
Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis.J Clin Microbiol. 2004 Aug;42(8):3462-8. doi: 10.1128/JCM.42.8.3462-3468.2004. J Clin Microbiol. 2004. PMID: 15297484 Free PMC article.
-
Feline bartonellosis and cat scratch disease.Vet Immunol Immunopathol. 2008 May 15;123(1-2):167-71. doi: 10.1016/j.vetimm.2008.01.025. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18295347 Review.
-
[Bartonella infection].Rev Med Chir Soc Med Nat Iasi. 2001 Jul-Sep;105(3):486-9. Rev Med Chir Soc Med Nat Iasi. 2001. PMID: 12092178 Review. Romanian.
Cited by
-
Optimization of pulse-field gel electrophoresis for subtyping of Klebsiella pneumoniae.Int J Environ Res Public Health. 2013 Jul 1;10(7):2720-31. doi: 10.3390/ijerph10072720. Int J Environ Res Public Health. 2013. PMID: 23880721 Free PMC article.
-
Evolutionary insights into scleractinian corals using comparative genomic hybridizations.BMC Genomics. 2012 Sep 21;13:501. doi: 10.1186/1471-2164-13-501. BMC Genomics. 2012. PMID: 22994626 Free PMC article.
-
The BatR/BatS two-component regulatory system controls the adaptive response of Bartonella henselae during human endothelial cell infection.J Bacteriol. 2010 Jul;192(13):3352-67. doi: 10.1128/JB.01676-09. Epub 2010 Apr 23. J Bacteriol. 2010. PMID: 20418395 Free PMC article.
-
A genome-wide study of recombination rate variation in Bartonella henselae.BMC Evol Biol. 2012 May 11;12:65. doi: 10.1186/1471-2148-12-65. BMC Evol Biol. 2012. PMID: 22577862 Free PMC article.
-
Genome dynamics of Bartonella grahamii in micro-populations of woodland rodents.BMC Genomics. 2010 Mar 4;11:152. doi: 10.1186/1471-2164-11-152. BMC Genomics. 2010. PMID: 20202191 Free PMC article.
References
-
- Alsmark, U. C. M., A. C. Frank, E. O. Karlberg, B. Legault, B. Canbäck, D. Ardell, A.-S. Eriksson, A. K. Näslund, S. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. E. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. - PMC - PubMed
-
- Amiri, H., W. Davids, and S. G. E. Andersson. 2003. Birth and death of orphan genes in Rickettsia. Mol. Biol. Evol. 20:1575-1587. - PubMed
-
- Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. M. Alsmark, R. M. Podowski, A. K. Näslund, A.-C. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. - PubMed
-
- Andersson, J. O., and S. G. E. Andersson. 1999. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16:1178-1191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

