Prediction of transcriptional terminators in Bacillus subtilis and related species
- PMID: 16110342
- PMCID: PMC1187862
- DOI: 10.1371/journal.pcbi.0010025
Prediction of transcriptional terminators in Bacillus subtilis and related species
Abstract
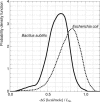
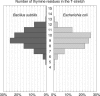
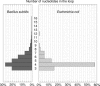
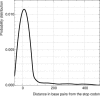
In prokaryotes, genes belonging to the same operon are transcribed in a single mRNA molecule. Transcription starts as the RNA polymerase binds to the promoter and continues until it reaches a transcriptional terminator. Some terminators rely on the presence of the Rho protein, whereas others function independently of Rho. Such Rho-independent terminators consist of an inverted repeat followed by a stretch of thymine residues, allowing us to predict their presence directly from the DNA sequence. Unlike in Escherichia coli, the Rho protein is dispensable in Bacillus subtilis, suggesting a limited role for Rho-dependent termination in this organism and possibly in other Firmicutes. We analyzed 463 experimentally known terminating sequences in B. subtilis and found a decision rule to distinguish Rho-independent transcriptional terminators from non-terminating sequences. The decision rule allowed us to find the boundaries of operons in B. subtilis with a sensitivity and specificity of about 94%. Using the same decision rule, we found an average sensitivity of 94% for 57 bacteria belonging to the Firmicutes phylum, and a considerably lower sensitivity for other bacteria. Our analysis shows that Rho-independent termination is dominant for Firmicutes in general, and that the properties of the transcriptional terminators are conserved. Terminator prediction can be used to reliably predict the operon structure in these organisms, even in the absence of experimentally known operons. Genome-wide predictions of Rho-independent terminators for the 57 Firmicutes are available in the Supporting Information section.
Conflict of interest statement
Figures


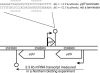

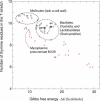
References
-
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18:S329–S336. - PubMed
-
- Makita Y, Nakao M, Ogasawara N, Nakai K. DBTBS: Database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:D75–D77. Available: http://dbtbs.hgc.jp. Accessed 12 July 2005. - PMC - PubMed
-
- Yada T, Nakao M, Totoki Y, Nakai K. Modeling and predicting transcriptional units of Escherichia coli genes using hidden Markov models. Bioinformatics. 1999;15:987–993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

