Comparative genomics and disorder prediction identify biologically relevant SH3 protein interactions
- PMID: 16110343
- PMCID: PMC1187863
- DOI: 10.1371/journal.pcbi.0010026
Comparative genomics and disorder prediction identify biologically relevant SH3 protein interactions
Abstract
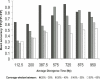
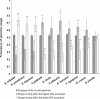
Protein interaction networks are an important part of the post-genomic effort to integrate a part-list view of the cell into system-level understanding. Using a set of 11 yeast genomes we show that combining comparative genomics and secondary structure information greatly increases consensus-based prediction of SH3 targets. Benchmarking of our method against positive and negative standards gave 83% accuracy with 26% coverage. The concept of an optimal divergence time for effective comparative genomics studies was analyzed, demonstrating that genomes of species that diverged very recently from Saccharomyces cerevisiae(S. mikatae, S. bayanus, and S. paradoxus), or a long time ago (Neurospora crassa and Schizosaccharomyces pombe), contain less information for accurate prediction of SH3 targets than species within the optimal divergence time proposed. We also show here that intrinsically disordered SH3 domain targets are more probable sites of interaction than equivalent sites within ordered regions. Our findings highlight several novel S. cerevisiae SH3 protein interactions, the value of selection of optimal divergence times in comparative genomics studies, and the importance of intrinsic disorder for protein interactions. Based on our results we propose novel roles for the S. cerevisiae proteins Abp1p in endocytosis and Hse1p in endosome protein sorting.
Conflict of interest statement
Figures



References
-
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, et al. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. - PubMed
-
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. - PubMed
-
- Gaasterland T, Ragan MA. Microbial genescapes: phyletic and functional patterns of ORF distribution among prokaryotes. Microb Comp Genomics. 1998;3:199–217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

