Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis
- PMID: 16111943
- PMCID: PMC5127709
- DOI: 10.1016/j.cub.2005.07.029
Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis
Abstract
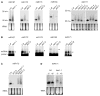
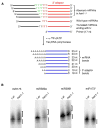
Small RNAs of 21-25 nucleotides (nt), including small interfering RNAs (siRNAs) and microRNAs (miRNAs), act as guide RNAs to silence target-gene expression in a sequence-specific manner. In addition to a Dicer homolog, DCL1, the biogenesis of miRNAs in Arabidopsis requires another protein, HEN1. miRNAs are reduced in abundance and increased in size in hen1 mutants. We found that HEN1 is a miRNA methyltransferase that adds a methyl group to the 3'-most nucleotide of miRNAs, but the role of miRNA methylation was unknown. Here, we show that siRNAs from sense transgenes, hairpin transgenes, and transposons or repeat sequences, as well as a new class of siRNAs known as trans-acting siRNAs, are also methylated in vivo by HEN1. In addition, we show that the size increase of small RNAs in the hen1-1 mutant is due to the addition of one to five U residues to the 3' ends of the small RNAs. Therefore, a novel uridylation activity targets the 3' ends of unmethylated miRNAs and siRNAs in hen1 mutants. We conclude that 3'-end methylation is a common step in miRNA and siRNA metabolism and likely protects the 3' ends of the small RNAs from the uridylation activity.
Figures


References
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

