Functional conservation of PISTILLATA activity in a pea homolog lacking the PI motif
- PMID: 16113230
- PMCID: PMC1203367
- DOI: 10.1104/pp.104.057687
Functional conservation of PISTILLATA activity in a pea homolog lacking the PI motif
Abstract
Current understanding of floral development is mainly based on what we know from Arabidopsis (Arabidopsis thaliana) and Antirrhinum majus. However, we can learn more by comparing developmental mechanisms that may explain morphological differences between species. A good example comes from the analysis of genes controlling flower development in pea (Pisum sativum), a plant with more complex leaves and inflorescences than Arabidopsis and Antirrhinum, and a different floral ontogeny. The analysis of UNIFOLIATA (UNI) and STAMINA PISTILLOIDA (STP), the pea orthologs of LEAFY and UNUSUAL FLORAL ORGANS, has revealed a common link in the regulation of flower and leaf development not apparent in Arabidopsis. While the Arabidopsis genes mainly behave as key regulators of flower development, where they control the expression of B-function genes, UNI and STP also contribute to the development of the pea compound leaf. Here, we describe the characterization of P. sativum PISTILLATA (PsPI), a pea MADS-box gene homologous to B-function genes like PI and GLOBOSA (GLO), from Arabidopsis and Antirrhinum, respectively. PsPI encodes for an atypical PI-type polypeptide that lacks the highly conserved C-terminal PI motif. Nevertheless, constitutive expression of PsPI in tobacco (Nicotiana tabacum) and Arabidopsis shows that it can specifically replace the function of PI, being able to complement the strong pi-1 mutant. Accordingly, PsPI expression in pea flowers, which is dependent on STP, is identical to PI and GLO. Interestingly, PsPI is also transiently expressed in young leaves, suggesting a role of PsPI in pea leaf development, a possibility that fits with the established role of UNI and STP in the control of this process.
Figures

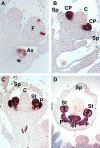


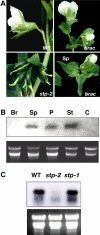

References
-
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199
-
- Berbel A, Navarro C, Ferrándiz C, Cañas LA, Madueño F, Beltrán JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25: 441–451 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

