Novel sialic acid transporter of Haemophilus influenzae
- PMID: 16113244
- PMCID: PMC1231074
- DOI: 10.1128/IAI.73.9.5291-5300.2005
Novel sialic acid transporter of Haemophilus influenzae
Abstract
Nontypeable Haemophilus influenzae is an opportunistic pathogen and a common cause of otitis media in children and of chronic bronchitis and pneumonia in patients with chronic obstructive pulmonary disease. The lipooligosaccharides, a major component of the outer membrane of H. influenzae, play an important role in microbial virulence and pathogenicity. N-Acetylneuraminic acid (sialic acid) can be incorporated into the lipooligosaccharides as a terminal nonreducing sugar. Although much of the pathway of sialic acid incorporation into lipooligosaccharides is understood, the transporter responsible for N-acetylneuraminic acid uptake in H. influenzae has yet to be characterized. In this paper we demonstrate that this transporter is a novel sugar transporter of the tripartite ATP-independent periplasmic transporter family. In the absence of this transporter, H. influenzae cannot incorporate sialic acid into its lipooligosaccharides, making the organism unable to survive when exposed to human serum and causing reduced viability in biofilm growth.
Figures
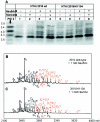

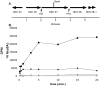
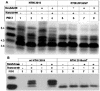


References
-
- Andreoni, J., H. Kayhty, and P. Densen. 1993. Vaccination and the role of capsular polysaccharide antibody in prevention of recurrent meningococcal disease in late complement component-deficient individuals. J. Infect. Dis. 168:227-231. - PubMed
-
- Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. - PubMed
-
- Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

