Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice
- PMID: 16113260
- PMCID: PMC1231100
- DOI: 10.1128/IAI.73.9.5438-5449.2005
Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice
Abstract
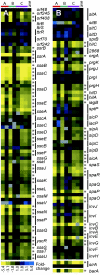
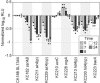
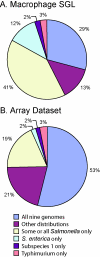
DNA microarrays provide an opportunity to combine the principles of signature-tagged mutagenesis (STM) with microarray technology to identify potentially important bacterial virulence genes. The scope of DNA microarrays allows for less laborious screening on a much larger scale than possible by STM alone. We have adapted a microarray-based transposon tracking strategy for use with a Salmonella enterica serovar Typhimurium cDNA microarray in order to identify genes important for survival and replication in RAW 264.7 mouse macrophage-like cells or in the spleens of BALB/cJ mice. A 50,000-CFU transposon library of S. enterica serovar Typhimurium strain SL1344 was serially passaged in cultured macrophages or intraperitoneally inoculated into BALB/cJ mice. The bacterial genomic DNA was isolated and processed for analysis on the microarray. The novel application of this approach to identify mutants unable to survive in cultured cells resulted in the identification of components of Salmonella pathogenicity island 2 (SPI2), which is known to be critical for intracellular survival and replication. In addition, array results indicated that a number of SPI1-associated genes, currently not associated with intracellular survival, are negatively selected. However, of the SPI1-associated mutants individually tested for intracellular survival, only a sirA mutant exhibited reduced numbers relative to those of wild-type bacteria. Of the mutants unable to survive in mice, significant proportions are either components of the SPI2 pathogenicity island or involved in lipopolysaccharide synthesis. This observation is in agreement with results obtained in the original S. enterica serovar Typhimurium STM screen, illustrating the utility of this approach for the high-throughput identification of virulence factors important for survival in the host.
Figures

References
-
- Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 160:59-62. - PubMed
-
- Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. - PubMed
-
- Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

