Genome-wide expression profiling in malaria infection reveals transcriptional changes associated with lethal and nonlethal outcomes
- PMID: 16113330
- PMCID: PMC1231079
- DOI: 10.1128/IAI.73.9.6091-6100.2005
Genome-wide expression profiling in malaria infection reveals transcriptional changes associated with lethal and nonlethal outcomes
Abstract
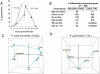
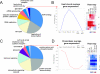
High-density oligonucleotide microarrays are widely used to study gene expression in cells exposed to a variety of pathogens. This study addressed the global genome-wide transcriptional activation of genes in hosts infected in vivo, which result in radically different clinical outcomes. We present an analysis of the gene expression profiles that identified a set of host biomarkers which distinguish between lethal and nonlethal blood stage Plasmodium yoelii malaria infections. Multiple biological replicates sampled during the course of infection were used to establish statistically valid sets of differentially expressed genes. These genes that correlated with the intensity of infection were used to identify pathways of cellular processes related to metabolic perturbations, erythropoiesis, and B-cell immune responses and other innate and cellular immune responses. The transcriptional apparatus that controls gene expression in erythropoiesis was also differentially expressed and regulated the expression of target genes involved in the host's response to malaria anemia. The biological systems approach provides unprecedented opportunities to explore the pathophysiology of host-pathogen interactions in experimental malaria infection and to decipher functionally complex networks of gene and protein interactions.
Figures




Similar articles
-
Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria.Infect Immun. 2006 Oct;74(10):5561-73. doi: 10.1128/IAI.00408-06. Infect Immun. 2006. PMID: 16988231 Free PMC article.
-
Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria.J Infect Dis. 2004 Apr 1;189(7):1245-56. doi: 10.1086/382596. Epub 2004 Mar 17. J Infect Dis. 2004. PMID: 15031794
-
Immunoregulation in murine malaria. Susceptibility of inbred mice to infection with Plasmodium yoelii depends on the dynamic interplay of host and parasite genes.J Immunol. 1988 Jul 1;141(1):241-8. J Immunol. 1988. PMID: 3379305
-
Microarrays for studying the host transcriptional response to microbial infection and for the identification of host drug targets.Microbes Infect. 2001 Aug;3(10):813-21. doi: 10.1016/s1286-4579(01)01439-3. Microbes Infect. 2001. PMID: 11580976 Review.
-
Cross-species comparison of genome-wide expression patterns.Genome Biol. 2004;5(7):232. doi: 10.1186/gb-2004-5-7-232. Epub 2004 Jun 21. Genome Biol. 2004. PMID: 15239823 Free PMC article. Review.
Cited by
-
Strain-specific innate immune signaling pathways determine malaria parasitemia dynamics and host mortality.Proc Natl Acad Sci U S A. 2014 Jan 28;111(4):E511-20. doi: 10.1073/pnas.1316467111. Epub 2014 Jan 13. Proc Natl Acad Sci U S A. 2014. PMID: 24474800 Free PMC article.
-
Differential gene expression mediated by 15-hydroxyeicosatetraenoic acid in LPS-stimulated RAW 264.7 cells.Malar J. 2009 Aug 11;8:195. doi: 10.1186/1475-2875-8-195. Malar J. 2009. PMID: 19671186 Free PMC article.
-
Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria.Infect Immun. 2006 Oct;74(10):5561-73. doi: 10.1128/IAI.00408-06. Infect Immun. 2006. PMID: 16988231 Free PMC article.
-
Comparative analysis of gene expression changes mediated by individual constituents of hemozoin.Chem Res Toxicol. 2009 Mar 16;22(3):433-45. doi: 10.1021/tx8002752. Chem Res Toxicol. 2009. PMID: 19191707 Free PMC article.
-
How "humane" is your endpoint? Refining the science-driven approach for termination of animal studies of chronic infection.PLoS Pathog. 2012 Jan;8(1):e1002399. doi: 10.1371/journal.ppat.1002399. Epub 2012 Jan 19. PLoS Pathog. 2012. PMID: 22275862 Free PMC article. No abstract available.
References
-
- Achtman, A. H., M. Khan, I. C. M. Maclennan, and J. Langhorne. 2003. Plasmodium chabaudi chabaudi infection in mice induces strong B-cell responses and striking but temporary changes in splenic cell distribution. J. Immunol. 171:317-324. - PubMed
-
- Alves, H. J., W. Weidanz, and L. Weiss. 1996. The spleen in murine Plasmodium chabaudi adami malaria: stromal cells, T lymphocytes, and hematopoiesis. Am. J. Trop. Med. Hyg. 55:370-378. - PubMed
-
- Andrews, N. C. 1994. Erythroid transcription factor NF-E2 coordinates hemoglobin synthesis. Ped. Res. 36:419-423. - PubMed
-
- Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

