Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro
- PMID: 16115957
- PMCID: PMC2171352
- DOI: 10.1083/jcb.200502073
Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro
Erratum in
- J Cell Biol. 2005 Sep 26;170(7):1169. Ramuns, John [corrected to Ramunas, John]
Abstract
The immortal strand hypothesis proposes that asymmetrically dividing stem cells (SCs) selectively segregate chromosomes that bear the oldest DNA templates. We investigated cosegregation in neural stem cells (NSCs). After exposure to the thymidine analogue 5-bromo-2-deoxyuridine (BrdU), which labels newly synthesized DNA, a subset of neural precursor cells were shown to retain BrdU signal. It was confirmed that some BrdU-retaining cells divided actively, and that these cells exhibited some characteristics of SCs. This asymmetric partitioning of DNA then was demonstrated during mitosis, and these results were further supported by real time imaging of SC clones, in which older and newly synthesized DNA templates were distributed asymmetrically after DNA synthesis. We demonstrate that NSCs are unique among precursor cells in the uneven partitioning of genetic material during cell divisions.
Figures

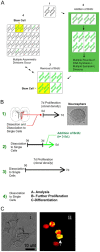

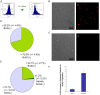
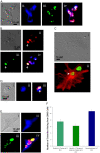
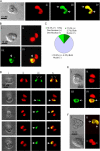

References
-
- Aguilaniu, H., L. Gustafsson, M. Rigoulet, and T. Nystrom. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 299:1751–1753. - PubMed
-
- Cairns, J. 1975. Mutation selection and the natural history of cancer. Nature. 255:197–200. - PubMed
-
- Dalgaard, J.Z., and A.J. Klar. 2001. Does S. pombe exploit the intrinsic asymmetry of DNA synthesis to imprint daughter cells for mating-type switching? Trends Genet. 17:153–157. - PubMed
-
- Doetsch, F., I. Caille, D.A. Lim, J.M. Garcia-Verdugo, and A. Alvarez-Buylla. 1999. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 97:703–716. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

