Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling
- PMID: 16117725
- PMCID: PMC1316262
- DOI: 10.1042/BJ20050873
Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling
Abstract
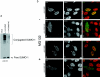
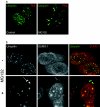
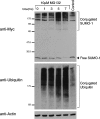
To investigate potential interplay between the SUMO1 (small ubiquitin-related modifier-1) and ubiquitin pathways of post-translational protein modification, we examined aspects of their localization and conjugation status during proteasome inhibition. Our results indicate that these pathways converge upon the discrete sub-nuclear domains known as PML (promyelocytic leukaemia protein) NBs (nuclear bodies). Proteasome inhibition generated an increased number of PML bodies, without any obvious increase in size. Using a cell line that constitutively expresses an epitope-tagged version of SUMO1, which was incorporated into high-molecular-mass conjugates, we observed SUMO1 accumulating in clusters around a subset of the NBs. Nuclear ubiquitin was initially observed in numerous speckles and foci, which bore no relationship to PML NBs in the absence of proteasome inhibition. However, during proteasome inhibition, total ubiquitin-conjugated species increased in the cell, as judged by Western blotting. Concomitantly the number of nuclear ubiquitin clusters decreased, and were almost quantitatively associated with the PML NBs, co-localizing with the SUMO-conjugated pool. Proteasome inhibition depleted the pool of free SUMO1 in the cell. Reversal of proteasome inhibition in the presence or absence of protein synthesis demonstrated that free SUMO1 was regenerated from the conjugated pool. The results indicate that a significant fraction of the free SUMO1 pool could be accounted for by recycling from the conjugated pool and indeed it may be that, as for ubiquitin, SUMO1 needs to be removed from conjugated species prior to processing by the proteasome. Taken together with other recent reports on the proteasome and PML NBs, these results suggest that the PML NBs may play an important role in integrating these pathways.
Figures





References
-
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. - PubMed
-
- Weissman A. M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. - PubMed
-
- Okura T., Gong L., Kamitani T., Wada T., Okura I., Wei C. F., Chang H. M., Yeh E. T. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. 1996;157:4277–4281. - PubMed
-
- Boddy M. N., Howe K., Etkin L. D., Solomon E., Freemont P. S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

