Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein
- PMID: 16120832
- PMCID: PMC1370824
- DOI: 10.1261/rna.2830905
Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein
Abstract
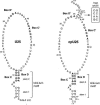
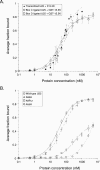
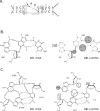
The interaction between box C/D small nucleolar (sno)RNAs and the 15.5K protein nucleates snoRNP assembly. Many eukaryotic snoRNAs contain two potential binding sites for this protein, only one of which appears to be utilized in vivo. The binding site conforms to the consensus for a kink-turn motif. We have investigated the molecular basis for selection of one potential site over the other using in vitro mobility shift assays and nucleotide analog interference mapping of Xenopus U25 snoRNA and of a circularly permuted form. We find that preferential binding of human 15.5K is not dependent on the proximity of RNA ends, but instead appears to require a structural context beyond the kink-turn itself. Direct analysis of the energetic contributions to binding made by 18 functional groups within the kink-turn identified both backbone atoms and base functionalities as key for interaction. An intramolecular RNA-RNA contact via a 2'-hydroxyl may supercede a putative Type I A-minor interaction in stabilizing the RNA-protein complex.
Figures



References
-
- Batey, R.T., Sagar, M.B., and Doudna, J.A. 2001. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 307: 229–246. - PubMed
-
- Bevilacqua, P.C. and Cech, T.R. 1996. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35: 9983–9994. - PubMed
-
- Cochrane, J.C. and Strobel, S.A. 2004. Probing RNA structure and function by nucleotide analog interference mapping. Curr. Protocols Nucleic Acid Chem. 6.9: 6.9.1–6.9.21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
