Local regulatory variation in Saccharomyces cerevisiae
- PMID: 16121257
- PMCID: PMC1189075
- DOI: 10.1371/journal.pgen.0010025
Local regulatory variation in Saccharomyces cerevisiae
Abstract
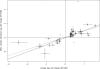
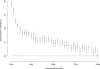
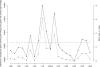
Naturally occurring sequence variation that affects gene expression is an important source of phenotypic differences among individuals within a species. We and others have previously shown that such regulatory variation can occur both at the same locus as the gene whose expression it affects (local regulatory variation) and elsewhere in the genome at trans-acting factors. Here we present a detailed analysis of genome-wide local regulatory variation in Saccharomyces cerevisiae. We used genetic linkage analysis to show that nearly a quarter of all yeast genes contain local regulatory variation between two divergent strains. We measured allele-specific expression in a diploid hybrid of the two strains for 77 genes showing strong self-linkage and found that in 52%-78% of these genes, local regulatory variation acts directly in cis. We also experimentally confirmed one example in which local regulatory variation in the gene AMN1 acts in trans through a feedback loop. Genome-wide sequence analysis revealed that genes subject to local regulatory variation show increased polymorphism in the promoter regions, and that some but not all of this increase is due to polymorphisms in predicted transcription factor binding sites. Increased polymorphism was also found in the 3' untranslated regions of these genes. These findings point to the importance of cis-acting variation, but also suggest that there is a diverse set of mechanisms through which local variation can affect gene expression levels.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
References
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
-
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. - PubMed
-
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics'. Nat Genet. 2005;37:225–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

